| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,088,110 – 5,088,210 |
| Length | 100 |
| Max. P | 0.698220 |

| Location | 5,088,110 – 5,088,210 |
|---|---|
| Length | 100 |
| Sequences | 4 |
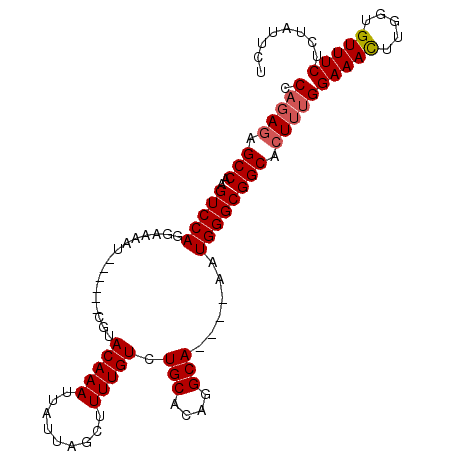
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 84.53 |
| Mean single sequence MFE | -27.18 |
| Consensus MFE | -19.33 |
| Energy contribution | -19.82 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5088110 100 + 22224390 CCAGAGAGCCAAGUCCAGGAAAAU------CGUACAAAUUAUUAGCUUUUGUCUGCACAGGCAAAUAAAUGGGCGGCACUUUGGAAACUUGGUGUUUCUCUAUUCU ..(((((((((((((((((....(------((..((..(((((.((((.((....)).)))).))))).))..)))...))))))..))))))...)))))..... ( -27.90) >DroSec_CAF1 5255 100 + 1 CCAGAGAGCCAAGUCCAGGAGAAU------CGUACAAAUUAUUAGCUUUUGUCUGCACAGGCAAGCAAAUGGGCGGCACUUUGGAAACUUGGUGUUUCUCUAUUCU ..((((((((((((((((.((..(------(((.....(((((.(((..(((((....)))))))).)))))))))..)))))))..))))))...)))))..... ( -28.30) >DroEre_CAF1 5356 89 + 1 CCAGAGAGCCAAGUCCAGAAAAAU------CCUACAAAUUAUAAGCUUUUGUCUGCCCAGGCA----AAUGGGCGGCACUUGGGAAAUUUGGUGUUUCU------- (((((...((((((((........------...(((((.........)))))..(((((....----..))))))).))))))....))))).......------- ( -25.60) >DroYak_CAF1 6122 102 + 1 CCAGAUAGCCAAGUCCAGGAAAACGUAAUACGUACAAAUUAUUAGCAUUUGUCUGCACAGGCA----AAUGGGCGGCACUAUGGAAAUUUGGUGUUUCUGUAUUUU .((((..(((((.((((.....(((.....)))............(((((((((....)))))----))))..........))))...)))))...))))...... ( -26.90) >consensus CCAGAGAGCCAAGUCCAGGAAAAU______CGUACAAAUUAUUAGCUUUUGUCUGCACAGGCA____AAUGGGCGGCACUUUGGAAACUUGGUGUUUCUCUAUUCU .(((((.(((..(((((................(((((.........))))).(((....)))......)))))))).)))))(((((.....)))))........ (-19.33 = -19.82 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:20:03 2006