
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,079,199 – 5,079,338 |
| Length | 139 |
| Max. P | 0.854844 |

| Location | 5,079,199 – 5,079,298 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 67.25 |
| Mean single sequence MFE | -19.93 |
| Consensus MFE | -16.09 |
| Energy contribution | -15.78 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.15 |
| Mean z-score | -0.73 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5079199 99 - 22224390 GAGAUGAGCAUCUGCAGCACGUCGUGCAGGUUCUCGUCCUGAAAGU--CAAACCAAAACGAGCGAUGAAAAACAGAAUGCCAAUUAAUUAAAAAGCAGUUA---- ..((((((.(((((((........))))))).))))))(((....(--((...............)))....)))..(((..............)))....---- ( -18.60) >DroVir_CAF1 54960 94 - 1 GAGAUGAGCAUCUGCAGCACAUCGUGCAGAUUCUCGUCCUGUGCAAAUGA---CA--AAAGGAGUUUCAAAU--AAAUAUAAUUUAAU----UAGCAGCCACUGU ((((((((.(((((((........))))))).))).((((.(((....).---))--..)))))))))....--..............----..((((...)))) ( -20.00) >DroPse_CAF1 39693 81 - 1 GAGAUGAGCAUUUGCAGCACAUCGUGCAGGUUCUCGUCCUGAAAAU--A----CGAAAUGCGAAGUCCAAAUC-CGAUAU-------------AGAACUCC---- ((..((..(.((((((.....(((((((((.......))))....)--)----)))..)))))))..))..))-......-------------........---- ( -16.20) >DroEre_CAF1 42651 99 - 1 GAGAUAAGCAUCUGCAGCACGUCGUGCAGAUUCUCGUCCUGAAAGU--CAAACCAAAACGAGGGGGGGAAAUCAUGAUGUUAAUUAAUUAAAUAGUUGUUA---- ..(((.((((((....((((...)))).(((((((.((((......--............)))).))).))))..)))))).)))................---- ( -20.37) >DroYak_CAF1 44518 96 - 1 GAGAUGAGCAUCUGCAGCACGUCAUGCAGAUUCUCGUCCUGGAAGU--UAAACCAAAACGAGG---GCAAAACAUGAUGUCAAUCAAUUAAAUAGCGGCUA---- ..((((((.(((((((........))))))).)))))).(((..((--((..((.......))---........((((....))))......))))..)))---- ( -24.60) >DroPer_CAF1 39472 81 - 1 GAGAUGAGCAUUUGCAGCACAUCGUGCAGGUUCUCGUCCUGAAAAU--A----CGAAAUGCGAAGUGCAAAUC-CGAUAU-------------AGAACUCC---- (.(((..((((((...(((..(((((((((.......))))....)--)----)))..))).))))))..)))-).....-------------........---- ( -19.80) >consensus GAGAUGAGCAUCUGCAGCACAUCGUGCAGAUUCUCGUCCUGAAAAU__AA___CAAAACGAGAGGUGCAAAUC_CGAUAU_AAUUAAU____UAGCAGUCA____ ..((((((.(((((((........))))))).))))))................................................................... (-16.09 = -15.78 + -0.30)



| Location | 5,079,229 – 5,079,338 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.70 |
| Mean single sequence MFE | -30.78 |
| Consensus MFE | -21.54 |
| Energy contribution | -22.07 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5079229 109 + 22224390 UUUUCAUCGCUCGUUUUGGUUUG--ACUUUCAGGACGAGAACCUGCACGACGUGCUGCAGAUGCUCAUCUCGCUGAUGUCGGAGCAUCCCAGCUCCAUGGUACCUGCCUUU .........((((((((((....--....)))))))))).....((((...)))).((((.((((((((.....))))..(((((......)))))..)))).)))).... ( -35.20) >DroVir_CAF1 54988 106 + 1 UUUGAAACUCCUUU--UG---UCAUUUGCACAGGACGAGAAUCUGCACGAUGUGCUGCAGAUGCUCAUCUCGUUGAUGUCGGAGCAUCCGAGCUCAAUGGUACCUGCCUUC ..............--..---.........(((((((((.(((((((........))))))).)))....((((((..((((.....))))..)))))))).))))..... ( -31.30) >DroPse_CAF1 39709 105 + 1 UUUGGACUUCGCAUUUCG----U--AUUUUCAGGACGAGAACCUGCACGAUGUGCUGCAAAUGCUCAUCUCAUUGAUGUCGGAGCAUCCCAGCUCCAUGGUACCUGCCUUC ....(((.((((((((((----(--.........))))))...)))..((((.((.......)).)))).....)).)))(((((......)))))..(((....)))... ( -29.20) >DroGri_CAF1 23956 101 + 1 UAUAUAUAUCUUGU--U--------CUGUUUAGGACGAGAAUCUGCACGAUGUUCUGCAGAUGCUCAUCUCACUGAUGUCGGAGCACCCGAGCUCAAUGGUACCUGCCUUC ........((((((--(--------((....)))))))))(((((((.(.....))))))))((.((((.....))))((((.....)))))).....(((....)))... ( -28.50) >DroAna_CAF1 16534 103 + 1 CUUGC---UCC-----UGCUCUUACUCUUACAGGACGAGAACCUGCACGACGUCCUGCAGAUGCUCAUCUCCCUGAUGUCGGAGCACCCCAGCUCCAUGGUACCUGCCUUC ((((.---(((-----((............))))))))).(((.....((((((..(.((((....)))).)..))))))(((((......)))))..))).......... ( -31.00) >DroPer_CAF1 39488 105 + 1 UUUGCACUUCGCAUUUCG----U--AUUUUCAGGACGAGAACCUGCACGAUGUGCUGCAAAUGCUCAUCUCAUUGAUGUCGGAGCAUCCCAGCUCCAUGGUACCUGCCUUC ((((((....((((.(((----(--.....((((.......)))).)))).))))))))))....((((.....))))..(((((......)))))..(((....)))... ( -29.50) >consensus UUUGCACCUCCCAU__UG____U__AUUUUCAGGACGAGAACCUGCACGAUGUGCUGCAGAUGCUCAUCUCACUGAUGUCGGAGCAUCCCAGCUCCAUGGUACCUGCCUUC ..............................(((((((((...(((((........)))))...)))..............(((((......)))))...)).))))..... (-21.54 = -22.07 + 0.53)

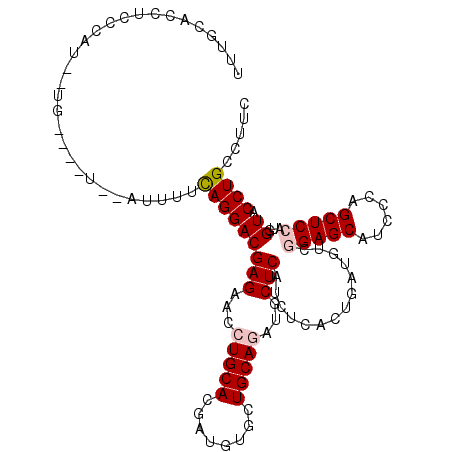

| Location | 5,079,229 – 5,079,338 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 79.70 |
| Mean single sequence MFE | -32.34 |
| Consensus MFE | -26.03 |
| Energy contribution | -26.08 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5079229 109 - 22224390 AAAGGCAGGUACCAUGGAGCUGGGAUGCUCCGACAUCAGCGAGAUGAGCAUCUGCAGCACGUCGUGCAGGUUCUCGUCCUGAAAGU--CAAACCAAAACGAGCGAUGAAAA .......(((.....(((((......)))))(((.((((...((((((.(((((((........))))))).))))))))))..))--)..)))................. ( -35.40) >DroVir_CAF1 54988 106 - 1 GAAGGCAGGUACCAUUGAGCUCGGAUGCUCCGACAUCAACGAGAUGAGCAUCUGCAGCACAUCGUGCAGAUUCUCGUCCUGUGCAAAUGA---CA--AAAGGAGUUUCAAA ...((......)).((((((((.((((......)))).(((.((((((.(((((((........))))))).)))))).)))........---..--....))).))))). ( -30.20) >DroPse_CAF1 39709 105 - 1 GAAGGCAGGUACCAUGGAGCUGGGAUGCUCCGACAUCAAUGAGAUGAGCAUUUGCAGCACAUCGUGCAGGUUCUCGUCCUGAAAAU--A----CGAAAUGCGAAGUCCAAA ....((((((....((((((......)))))).((((.....))))...)))))).(((..(((((((((.......))))....)--)----)))..))).......... ( -30.40) >DroGri_CAF1 23956 101 - 1 GAAGGCAGGUACCAUUGAGCUCGGGUGCUCCGACAUCAGUGAGAUGAGCAUCUGCAGAACAUCGUGCAGAUUCUCGUCCUAAACAG--------A--ACAAGAUAUAUAUA ............((((((..((((.....))))..)))))).((((((.((((((((.....).))))))).))))))........--------.--.............. ( -29.10) >DroAna_CAF1 16534 103 - 1 GAAGGCAGGUACCAUGGAGCUGGGGUGCUCCGACAUCAGGGAGAUGAGCAUCUGCAGGACGUCGUGCAGGUUCUCGUCCUGUAAGAGUAAGAGCA-----GGA---GCAAG ((..((..((((...(((((......)))))(((.((..(.((((....)))).)..)).)))))))..))..)).((((((..........)))-----)))---..... ( -36.90) >DroPer_CAF1 39488 105 - 1 GAAGGCAGGUACCAUGGAGCUGGGAUGCUCCGACAUCAAUGAGAUGAGCAUUUGCAGCACAUCGUGCAGGUUCUCGUCCUGAAAAU--A----CGAAAUGCGAAGUGCAAA ....((((((....((((((......)))))).((((.....))))...)))))).((((.((((.((((.......)))).....--.----......)))).))))... ( -32.02) >consensus GAAGGCAGGUACCAUGGAGCUGGGAUGCUCCGACAUCAAUGAGAUGAGCAUCUGCAGCACAUCGUGCAGGUUCUCGUCCUGAAAAU__A____CA__ACGAGAAAUGCAAA .....(((......((((((......))))))..........((((((.(((((((........))))))).))))))))).............................. (-26.03 = -26.08 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:59 2006