
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,069,102 – 5,069,232 |
| Length | 130 |
| Max. P | 0.938072 |

| Location | 5,069,102 – 5,069,192 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 81.23 |
| Mean single sequence MFE | -22.82 |
| Consensus MFE | -15.44 |
| Energy contribution | -15.83 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5069102 90 - 22224390 CAUAGGGCCAUUUGGCCAGUGGCGGCAGUACCAUGGCCUGUACGAAUGUAAAACGGAAACGGAAAUGAGACCGAAAAGCGAUUAAA-AGCA ....((.(((((((((((.(((........))))))))...............((....)).))))).).)).....((.......-.)). ( -22.70) >DroSec_CAF1 26822 87 - 1 CAUAGGGCCAUUUGGCCAGCGGCGGCAGGACCAUGGCCUGUACGAGCG---AACGAAAACGGAAAUGAGACCGAAAAACGAUUACA-AGCA .....((((....)))).((.((.(((((.(....))))))....)).---........(((........))).............-.)). ( -23.50) >DroSim_CAF1 26985 87 - 1 CAUAGGGCCAUUUGGCCAGCGGCGGCAGGACCAUGGCCUGUACGAGCG---AACGAAAACGGAAAUGAGACCGAAAAACGAUUACA-AGCA .....((((....)))).((.((.(((((.(....))))))....)).---........(((........))).............-.)). ( -23.50) >DroEre_CAF1 32846 83 - 1 CAUAUGGCCAUUUGGCCAGUGGCGGCAGGACCAUGGCCUGUACG-------AACGAAAACGGAAAUGAUACCGGAAAACGAUUACA-ACCA ....(((((....)))))(((((((((((.(....))))))...-------........(((........))).....)).)))).-.... ( -23.30) >DroWil_CAF1 7982 82 - 1 CAUAAGGCCAUUUUGCCAACGGUGGCAGGACCAUGGCCUAGAAAAAAA---AACAAUAGAGCAAAA---GCC--AAAGCAAUUAUA-CACA ....((((((((((((((....))))))))...)))))).........---.......(.((....---)))--............-.... ( -19.20) >DroYak_CAF1 34651 84 - 1 CAUAGGGCCAUUUGGCCAGUGGCGGCAGGACCAUGGCCUGCACG-------AACGAAAACGGAAAUGAAGCCGAAAAACGAUUACAAACUA ..(((((((....)))).(((((((((((.(....))))))...-------........(((........))).....)).))))...))) ( -24.70) >consensus CAUAGGGCCAUUUGGCCAGCGGCGGCAGGACCAUGGCCUGUACGA__G___AACGAAAACGGAAAUGAGACCGAAAAACGAUUACA_AGCA ....(((((((...(((......)))......)))))))....................(((........))).................. (-15.44 = -15.83 + 0.39)

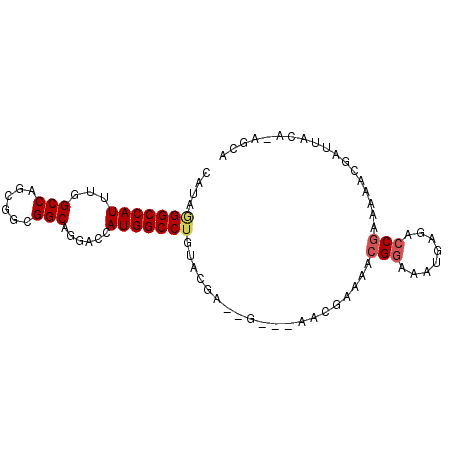
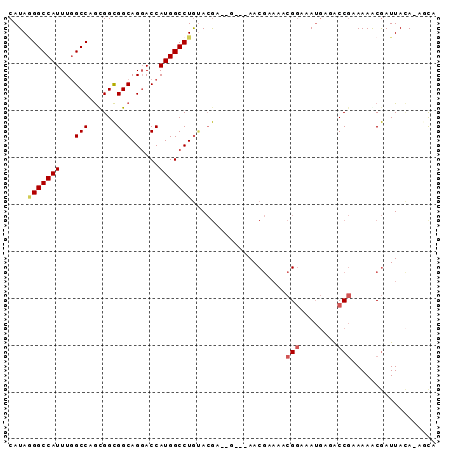
| Location | 5,069,130 – 5,069,232 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 74.97 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -20.77 |
| Energy contribution | -20.63 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5069130 102 + 22224390 CCGUUUCCGUUUUACAUUCGUACAGGCCAUGGUACUGCCGCCACUGGCCAAAUGGCCCUAUGAGAACGGAUUCACCUUUACCACCUGGUUCCGCCUGGAUCC ..(..((((((((.(((..(..(((((((((((......)))).)))))...))..)..)))))))))))..)..........((.((.....)).)).... ( -31.30) >DroGri_CAF1 9216 90 + 1 CCGCAU---U---------CCACAGGCAAUGGUUUUGCCACCGUUGGCCAAAUGGCCCUACGAAAAUGGAUUUACCUUUACCACCUGGUUUCGAUUGGAUCC ......---.---------.....(((((.....)))))..(((.((((....))))..))).....(((((((.....(((....)))......))))))) ( -24.70) >DroWil_CAF1 8005 99 + 1 GCUCUAUUGUU---UUUUUUUCUAGGCCAUGGUCCUGCCACCGUUGGCAAAAUGGCCUUAUGAGAAUGGAUUUACCUUUACCACAUGGUUUCGAUUGGAUCC ...........---...(((((.(((((((.....(((((....)))))..)))))))...))))).(((((((.....(((....)))......))))))) ( -27.80) >DroMoj_CAF1 28286 84 + 1 ---------U---------CCACAGGCAAUGGUGCUGCCGCCGCUGGCCAAGUGGCCAUACGAGAAUGGAUUCACCUUCACCACCUGGUUUCGAUUGGAUCC ---------(---------(((..((((.......))))((((((.....))))))....((((((.((.....)))))(((....))).)))..))))... ( -24.20) >DroAna_CAF1 5477 85 + 1 -----------------UGUUGCAGGCCAUGGUCCUGCCUCCCCUGGCAAAGUGGCCUUACGAGAACGGAUUCACCUUCACCACCUGGUUUCGGCUGGAUCC -----------------.((...(((((((.....((((......))))..))))))).))......(((((((((...(((....)))...)).))))))) ( -30.20) >DroPer_CAF1 29828 93 + 1 CCGUAUUUAUU---------UGCAGGCCAUGGUCCUGCCGCCGCUGGCCAAAUGGCCGUAUGAGAAUGGAUUUACCUUUACCACAUGGUUUCGCCUGGAUCC .(((((.....---------.((((((....).))))).......((((....))))))))).....(((((((.....(((....)))......))))))) ( -26.20) >consensus CCG__U___U_________CUACAGGCCAUGGUCCUGCCGCCGCUGGCCAAAUGGCCCUACGAGAAUGGAUUCACCUUUACCACCUGGUUUCGACUGGAUCC ........................((((((......(((......)))...))))))..........(((((((.....(((....)))......))))))) (-20.77 = -20.63 + -0.14)


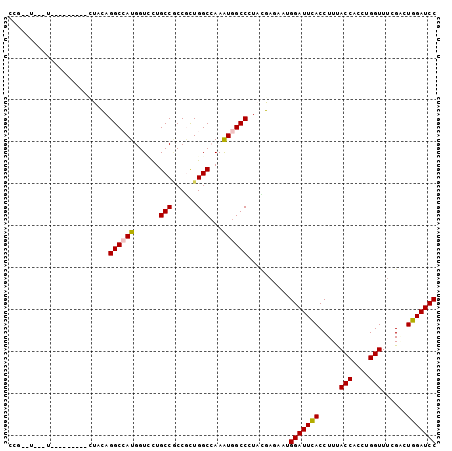
| Location | 5,069,130 – 5,069,232 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 74.97 |
| Mean single sequence MFE | -28.87 |
| Consensus MFE | -18.91 |
| Energy contribution | -19.80 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5069130 102 - 22224390 GGAUCCAGGCGGAACCAGGUGGUAAAGGUGAAUCCGUUCUCAUAGGGCCAUUUGGCCAGUGGCGGCAGUACCAUGGCCUGUACGAAUGUAAAACGGAAACGG .....(((((...(((....)))...((((...((((...(((..((((....)))).)))))))...))))...))))).............((....)). ( -33.70) >DroGri_CAF1 9216 90 - 1 GGAUCCAAUCGAAACCAGGUGGUAAAGGUAAAUCCAUUUUCGUAGGGCCAUUUGGCCAACGGUGGCAAAACCAUUGCCUGUGG---------A---AUGCGG .........((...(((.(.(((((.(((....((((...(((..((((....)))).)))))))....))).)))))).)))---------.---...)). ( -26.90) >DroWil_CAF1 8005 99 - 1 GGAUCCAAUCGAAACCAUGUGGUAAAGGUAAAUCCAUUCUCAUAAGGCCAUUUUGCCAACGGUGGCAGGACCAUGGCCUAGAAAAAAA---AACAAUAGAGC ((((...(((...(((....)))...)))..)))).((((....((((((((((((((....))))))))...)))))))))).....---........... ( -27.20) >DroMoj_CAF1 28286 84 - 1 GGAUCCAAUCGAAACCAGGUGGUGAAGGUGAAUCCAUUCUCGUAUGGCCACUUGGCCAGCGGCGGCAGCACCAUUGCCUGUGG---------A--------- ..............(((.(.((..(.((((...((....((((.(((((....))))))))).))...)))).)..))).)))---------.--------- ( -29.30) >DroAna_CAF1 5477 85 - 1 GGAUCCAGCCGAAACCAGGUGGUGAAGGUGAAUCCGUUCUCGUAAGGCCACUUUGCCAGGGGAGGCAGGACCAUGGCCUGCAACA----------------- ((((...(((...(((....)))...)))..))))((((((.(..(((......)))..).)))(((((.(....))))))))).----------------- ( -30.80) >DroPer_CAF1 29828 93 - 1 GGAUCCAGGCGAAACCAUGUGGUAAAGGUAAAUCCAUUCUCAUACGGCCAUUUGGCCAGCGGCGGCAGGACCAUGGCCUGCA---------AAUAAAUACGG ....((.(.((......((((..((.((.....)).))..)))).((((....))))..)).).(((((.(....)))))).---------.........)) ( -25.30) >consensus GGAUCCAAUCGAAACCAGGUGGUAAAGGUAAAUCCAUUCUCAUAAGGCCAUUUGGCCAGCGGCGGCAGGACCAUGGCCUGUAA_________A___A__CGG ((((...(((...(((....)))...)))..)))).........(((((((...(((......)))......)))))))....................... (-18.91 = -19.80 + 0.89)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:52 2006