| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 645,257 – 645,365 |
| Length | 108 |
| Max. P | 0.796032 |

| Location | 645,257 – 645,365 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.39 |
| Mean single sequence MFE | -33.27 |
| Consensus MFE | -24.86 |
| Energy contribution | -26.42 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
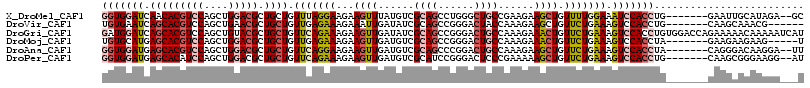
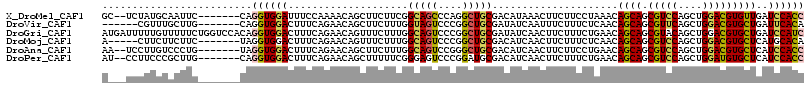
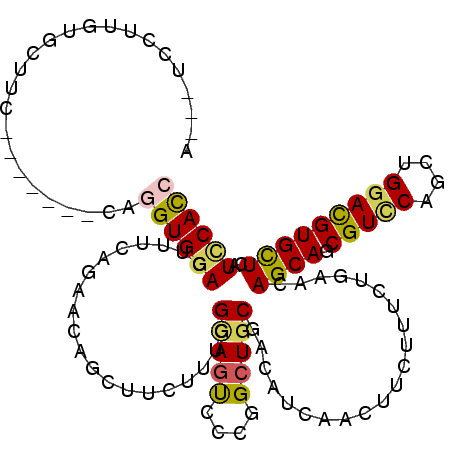
>X_DroMel_CAF1 645257 108 + 22224390 GGUGGAUCAACACGUCCAGCUGGACGCUGCUGUUUAGGAAGAAGUUUAUGUCGCAGCCUGGGCUGCCGAAGAAGCUGUUUUGGAAAUCCACCUG-------GAAUUGCAUAGA--GC (((((((((.(((((((....))))).)).))(((((((...(((((...(((((((....)))).)))..))))).))))))).)))))))..-------............--.. ( -37.30) >DroVir_CAF1 5129 104 + 1 UGUGAAUCAGCACGUCCAGCUGAACGCUGCUGUUGAGAAAGAAAUUGAUAUCGCAGCCGGGACUACCAAAGAAGCUGUUCUGAAAGUCCACCUG-------CAAGCAAACG------ .(((..(((((.......))))).)))((((...((..((....))....))((((...(((((.....((((....))))...)))))..)))-------).))))....------ ( -26.50) >DroGri_CAF1 5093 117 + 1 GAUGGAUCAGCACGUCCAGCUGUACGCUGCUGUUCAGAAAGAAGUUGAUAUCGCAGCCGGGACUGCCAAAGAAACUGUUCUGAAAGUCCACCUGUGGACCAGAAAAACAAAAAUCAU (((....((((((((.(....).))).)))))(((((((...((((......((((......))))......)))).))))))).(((((....))))).............))).. ( -31.70) >DroMoj_CAF1 5584 105 + 1 UGUGCAUGAGCACGUCCAGCUGGACGCUGCUGUUGAGAAAGAAGUUGAUGUCGCAGCCGGGACUGCCAAAGAAACUGUUCUGAAAGUCCACCUA-------GAAGAAGAAG-----U .((((....))))((((....))))(((((.((..(........)..))...)))))..(((((.....((((....))))...))))).....-------..........-----. ( -28.30) >DroAna_CAF1 3509 108 + 1 GGUGGAUGAGCACGUCCAGCUGGACGCUGCUGUUCAGGAAGAAGUUGAUGUCGCAGCCCGGACUGCCAAAGAAGCUGUUCUGAAAGUCCACCUA-------CAGGGACAAGGA--UU (((((((.(((((((((....))))).)))).(((((((...((((..(...((((......))))...)..)))).))))))).)))))))..-------............--.. ( -40.20) >DroPer_CAF1 14933 108 + 1 GGUGGAUGAGCACAUCCAGCUGGACGCUGCUGUUCAGAAAGAAGUUGAUGUCGCAUCCGGGACUCCCGAAAAAGCUGUUCUGAAAGUCCACCUG-------CAAGCGGGAAGG--AU (.((((((....)))))).)(((((.......(((((((...((((((((...))))((((...))))....)))).))))))).)))))((((-------....))))....--.. ( -35.60) >consensus GGUGGAUCAGCACGUCCAGCUGGACGCUGCUGUUCAGAAAGAAGUUGAUGUCGCAGCCGGGACUGCCAAAGAAGCUGUUCUGAAAGUCCACCUG_______CAAGAACAAAGA___U (((((((.(((((((((....))))).)))).(((((((...((((......((((......))))......)))).))))))).)))))))......................... (-24.86 = -26.42 + 1.56)
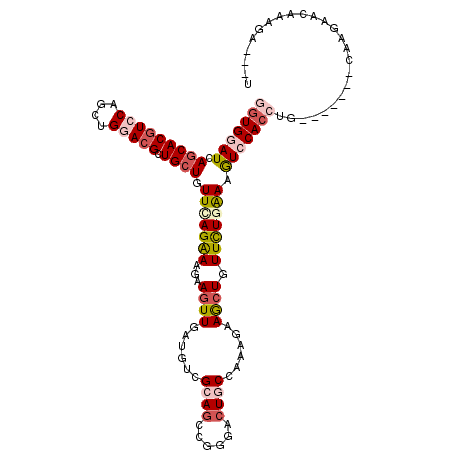
| Location | 645,257 – 645,365 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.39 |
| Mean single sequence MFE | -32.95 |
| Consensus MFE | -22.70 |
| Energy contribution | -22.82 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 645257 108 - 22224390 GC--UCUAUGCAAUUC-------CAGGUGGAUUUCCAAAACAGCUUCUUCGGCAGCCCAGGCUGCGACAUAAACUUCUUCCUAAACAGCAGCGUCCAGCUGGACGUGUUGAUCCACC ((--.....)).....-------..(((((((...................(((((....)))))....................(((((.(((((....))))))))))))))))) ( -35.30) >DroVir_CAF1 5129 104 - 1 ------CGUUUGCUUG-------CAGGUGGACUUUCAGAACAGCUUCUUUGGUAGUCCCGGCUGCGAUAUCAAUUUCUUUCUCAACAGCAGCGUUCAGCUGGACGUGCUGAUUCACA ------.......(((-------(((.((((((.(((((........))))).))).))).))))))..................(((((.(((((....))))))))))....... ( -31.10) >DroGri_CAF1 5093 117 - 1 AUGAUUUUUGUUUUUCUGGUCCACAGGUGGACUUUCAGAACAGUUUCUUUGGCAGUCCCGGCUGCGAUAUCAACUUCUUUCUGAACAGCAGCGUACAGCUGGACGUGCUGAUCCAUC ..((((..((((...((((((((....))))).(((((((.((((....(.(((((....))))).)....))))...))))))))))((((.....))))))))....)))).... ( -33.90) >DroMoj_CAF1 5584 105 - 1 A-----CUUCUUCUUC-------UAGGUGGACUUUCAGAACAGUUUCUUUGGCAGUCCCGGCUGCGACAUCAACUUCUUUCUCAACAGCAGCGUCCAGCUGGACGUGCUCAUGCACA .-----........((-------(((.(((((...((((........))))(((((....)))))...........................))))).))))).((((....)))). ( -30.00) >DroAna_CAF1 3509 108 - 1 AA--UCCUUGUCCCUG-------UAGGUGGACUUUCAGAACAGCUUCUUUGGCAGUCCGGGCUGCGACAUCAACUUCUUCCUGAACAGCAGCGUCCAGCUGGACGUGCUCAUCCACC ..--............-------..((((((..(((((..(((.....)))(((((....)))))...............))))).((((.(((((....)))))))))..)))))) ( -33.10) >DroPer_CAF1 14933 108 - 1 AU--CCUUCCCGCUUG-------CAGGUGGACUUUCAGAACAGCUUUUUCGGGAGUCCCGGAUGCGACAUCAACUUCUUUCUGAACAGCAGCGUCCAGCUGGAUGUGCUCAUCCACC ..--............-------..((((((..(((((((..((...((((((...)))))).))(....).......))))))).((((.(((((....)))))))))..)))))) ( -34.30) >consensus A___UCCUUGUGCUUC_______CAGGUGGACUUUCAGAACAGCUUCUUUGGCAGUCCCGGCUGCGACAUCAACUUCUUUCUGAACAGCAGCGUCCAGCUGGACGUGCUCAUCCACC .........................((((((....................(((((....))))).....................((((.(((((....)))))))))..)))))) (-22.70 = -22.82 + 0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:22 2006