
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,053,149 – 5,053,248 |
| Length | 99 |
| Max. P | 0.821804 |

| Location | 5,053,149 – 5,053,248 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.07 |
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -22.66 |
| Energy contribution | -23.22 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
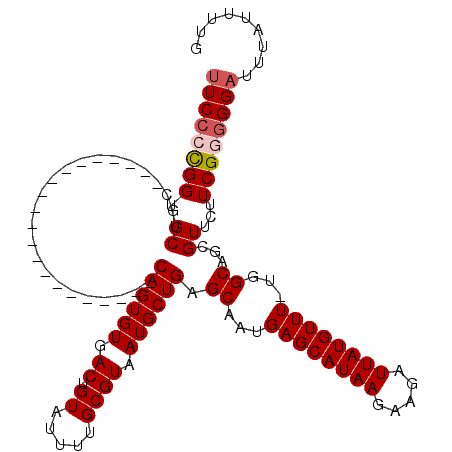
>X_DroMel_CAF1 5053149 99 + 22224390 UUCCCCGGUGCUGC---------------------CAGUGUGACUGUAUUUUGCGUAAUGCUGAGCAAUGAGCAUAAGAAGAUUAUGUUUUUCGCAACGUUCUUCGGGGGAUUUAUUUUG ((((((.(((((((---------------------((((((.((.((.....)))).)))))).))....)))))..(((((..(((((......))))))))))))))))......... ( -29.40) >DroSec_CAF1 11582 98 + 1 UUCCCCGGUGCUGC---------------------CAGUGUGACUGUAUUUUGCGUAAUGCUGAGCAAUGAGCAUAAGAAGAUUAUGUUU-UGGCAGCGUUCUUCGAGGGAUUUAUUUUG .(((((((((((((---------------------((.(((....(((((......)))))...)))..((((((((.....))))))))-))))))))....))).))))......... ( -31.30) >DroSim_CAF1 11823 98 + 1 UUCCCCGGUGCUGC---------------------CAGUGUGACUGUAUUUUGCGUAAUGCUGAGCAAUGAGCAUAAGAAGAUUAUGUUU-UGGCAGCGUUCUUCGAGGGAUUUAUUUUG .(((((((((((((---------------------((.(((....(((((......)))))...)))..((((((((.....))))))))-))))))))....))).))))......... ( -31.30) >DroEre_CAF1 17026 117 + 1 UUCCCUGGUGCUGCCUGUUUCCAUUCACCA--GGCCAGUGUGACUGUAUUUUGCGUAAUGCUGAGCAAUGAGCAUAAGAAGAUUAUGUUU-UGGCAACGUUCUUCGGGGGAUUUAUUUUA ((((((((.((.(((((...........))--)))((((((.((.((.....)))).)))))).((...((((((((.....))))))))-..))...))...))))))))......... ( -32.00) >DroYak_CAF1 19409 118 + 1 CUCCCUGGUGCUGCCUGUUUCCAGUUUCCAUUCGCCAGUGUGACUGUGUUUUGCGUAAUGCUGAGCAAUGAGCAUAAGAAGAUUAUGUUU-UGGCAGCGUUCUUCGG-GGAUUUAUUUUG .((((((((((((((.............((((.((((((((.((.((.....)))).)))))).))))))(((((((.....))))))).-.)))))))....))))-)))......... ( -37.10) >consensus UUCCCCGGUGCUGC_____________________CAGUGUGACUGUAUUUUGCGUAAUGCUGAGCAAUGAGCAUAAGAAGAUUAUGUUU_UGGCAGCGUUCUUCGGGGGAUUUAUUUUG ((((((((.((........................((((((.((.((.....)))).)))))).((...((((((((.....))))))))...))...))...))))))))......... (-22.66 = -23.22 + 0.56)

| Location | 5,053,149 – 5,053,248 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.07 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -16.58 |
| Energy contribution | -16.86 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5053149 99 - 22224390 CAAAAUAAAUCCCCCGAAGAACGUUGCGAAAAACAUAAUCUUCUUAUGCUCAUUGCUCAGCAUUACGCAAAAUACAGUCACACUG---------------------GCAGCACCGGGGAA .........(((((.(((((..(((......)))....)))))...(((((.((((..........))))....(((.....)))---------------------).))))..))))). ( -21.90) >DroSec_CAF1 11582 98 - 1 CAAAAUAAAUCCCUCGAAGAACGCUGCCA-AAACAUAAUCUUCUUAUGCUCAUUGCUCAGCAUUACGCAAAAUACAGUCACACUG---------------------GCAGCACCGGGGAA .........((((.((......(((((((-...(((((.....)))))....((((..........)))).............))---------------------)))))..)))))). ( -22.30) >DroSim_CAF1 11823 98 - 1 CAAAAUAAAUCCCUCGAAGAACGCUGCCA-AAACAUAAUCUUCUUAUGCUCAUUGCUCAGCAUUACGCAAAAUACAGUCACACUG---------------------GCAGCACCGGGGAA .........((((.((......(((((((-...(((((.....)))))....((((..........)))).............))---------------------)))))..)))))). ( -22.30) >DroEre_CAF1 17026 117 - 1 UAAAAUAAAUCCCCCGAAGAACGUUGCCA-AAACAUAAUCUUCUUAUGCUCAUUGCUCAGCAUUACGCAAAAUACAGUCACACUGGCC--UGGUGAAUGGAAACAGGCAGCACCAGGGAA ...............(((((..(((....-.)))....)))))...(((....(((...)))....))).....(((.....))).((--(((((..((....)).....)))))))... ( -26.70) >DroYak_CAF1 19409 118 - 1 CAAAAUAAAUCC-CCGAAGAACGCUGCCA-AAACAUAAUCUUCUUAUGCUCAUUGCUCAGCAUUACGCAAAACACAGUCACACUGGCGAAUGGAAACUGGAAACAGGCAGCACCAGGGAG .........(((-(.(.....)((((((.-...(((((.....))))).(((((((...((.....))......(((.....))))).)))))....((....))))))))....)))). ( -27.80) >consensus CAAAAUAAAUCCCCCGAAGAACGCUGCCA_AAACAUAAUCUUCUUAUGCUCAUUGCUCAGCAUUACGCAAAAUACAGUCACACUG_____________________GCAGCACCGGGGAA .........(((((.(.....)(((((...................(((....(((...)))....))).....(((.....))).....................)))))...))))). (-16.58 = -16.86 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:46 2006