
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,046,070 – 5,046,221 |
| Length | 151 |
| Max. P | 0.856920 |

| Location | 5,046,070 – 5,046,187 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.49 |
| Mean single sequence MFE | -24.34 |
| Consensus MFE | -14.07 |
| Energy contribution | -14.88 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5046070 117 - 22224390 UAUUAUAUAGCUAAAUUUCAGUGCGCGAAAGCCACAAGAAAUUCAACUAUUUAAAAUAUAAUAA-AGAAAAGUGGGUGGAAAAAAUAGAGGAGUGCUGUGGGGGAUGGGG-GGAUGGCA .........((((..(..(.((.(((....))((((....((((..((((((....(((.((..-......))..)))....))))))..))))..))))..).)).)..-)..)))). ( -22.50) >DroSec_CAF1 4958 119 - 1 UAUAAUAUAGCUAAAUUUCAGUGCGCGAAAGCCACAAGAAAUUCAUCUAUUUAAAACAUAAUAAAAGAAAAGUGGAUGGCGAAACUAGGGGACUGGUGUGUGGGGGGGGGGGAGUGGCA .........((((..(..(.(((.((....))))).......((((((((((.................))))))))))((..(((((....)))))...))......)..)..)))). ( -31.13) >DroSim_CAF1 4961 110 - 1 UAUAAUAUAGGUACAUUUCAGUGCGCGAAAGCCACAAGAAAUUCAUCUAUUUAAAACAUAAUAAAAGAAAAGUGGGUGGCGAAACUAGAGGAGUAUUGCG---------GGGGGUGGCA ..........((.(((..(.(((.((....)))))........(((((((((.................)))))))))((((.(((.....))).)))).---------.)..))))). ( -25.53) >DroYak_CAF1 10280 107 - 1 UGUUAUAUAGCGAAAUUCCAUUGCGCGAAAGCCACAAGAAAUUAAUCCAUUUAAAACAUAAUAAAAGA-AAGUGGAAAGAAAAAAUAGAGGAGGGUUGAGGGG----------G-UGCA .((....(..(((..((((..((.((....))))...........(((((((................-))))))).............))))..)))..)..----------.-.)). ( -18.19) >consensus UAUAAUAUAGCUAAAUUUCAGUGCGCGAAAGCCACAAGAAAUUCAUCUAUUUAAAACAUAAUAAAAGAAAAGUGGAUGGAAAAAAUAGAGGAGUGUUGUGGGG______G_GGGUGGCA .........((((.......(((.((....))))).......((((((((((.................))))))))))...................................)))). (-14.07 = -14.88 + 0.81)



| Location | 5,046,109 – 5,046,221 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 85.88 |
| Mean single sequence MFE | -17.69 |
| Consensus MFE | -11.69 |
| Energy contribution | -12.50 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5046109 112 + 22224390 CCACCCACUUUUCU-UUAUUAUAUUUUAAAUAGUUGAAUUUCUUGUGGCUUUCGCGCACUGAAAUUUAGCUAUAUAAUAAGUUUUUCAAACAUU---UUUUAGCUUAUUUAUAAUU ..............-..((((((......(((((((((((((..(((((....)).))).)))))))))))))..((((((((...........---....)))))))))))))). ( -22.66) >DroSec_CAF1 4998 113 + 1 CCAUCCACUUUUCUUUUAUUAUGUUUUAAAUAGAUGAAUUUCUUGUGGCUUUCGCGCACUGAAAUUUAGCUAUAUUAUAAGUUUUUCAUCCAUU---UUUUAGCCUAUUUAAAAUU ......................(((((((((((.((((((((..(((((....)).))).))))))))((((......................---...))))))))))))))). ( -20.61) >DroSim_CAF1 4992 113 + 1 CCACCCACUUUUCUUUUAUUAUGUUUUAAAUAGAUGAAUUUCUUGUGGCUUUCGCGCACUGAAAUGUACCUAUAUUAUAAGUUUUUCAUCCAUU---UUUUAGCCUAUUUAAAAUU .............(((((.((.(((..((((.((((((...((((((((....))(((......)))........))))))...)))))).)))---)...))).))..))))).. ( -14.90) >DroYak_CAF1 10309 115 + 1 CUUUCCACUU-UCUUUUAUUAUGUUUUAAAUGGAUUAAUUUCUUGUGGCUUUCGCGCAAUGGAAUUUCGCUAUAUAACAAGUUCUUCACCAAUUAAGUUUUAAAUUAUAUACGACU ..(((((..(-((.((((........)))).)))........(((((.......))))))))))..(((.(((((((..((..(((........)))..))...)))))))))).. ( -12.60) >consensus CCACCCACUUUUCUUUUAUUAUGUUUUAAAUAGAUGAAUUUCUUGUGGCUUUCGCGCACUGAAAUUUAGCUAUAUAAUAAGUUUUUCAUCCAUU___UUUUAGCCUAUUUAAAAUU ......................(((((((((((.((((((((..(((((....)).))).))))))))(((........)))......................))))))))))). (-11.69 = -12.50 + 0.81)

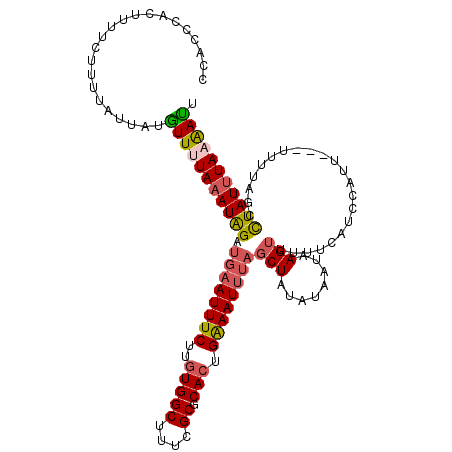

| Location | 5,046,109 – 5,046,221 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 85.88 |
| Mean single sequence MFE | -19.65 |
| Consensus MFE | -11.90 |
| Energy contribution | -12.40 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5046109 112 - 22224390 AAUUAUAAAUAAGCUAAAA---AAUGUUUGAAAAACUUAUUAUAUAGCUAAAUUUCAGUGCGCGAAAGCCACAAGAAAUUCAACUAUUUAAAAUAUAAUAA-AGAAAAGUGGGUGG .((((((..(((((.....---...))))).........(((.((((...((((((.(((.((....)))))..))))))...)))).)))..))))))..-.............. ( -17.90) >DroSec_CAF1 4998 113 - 1 AAUUUUAAAUAGGCUAAAA---AAUGGAUGAAAAACUUAUAAUAUAGCUAAAUUUCAGUGCGCGAAAGCCACAAGAAAUUCAUCUAUUUAAAACAUAAUAAAAGAAAAGUGGAUGG ...........(((((...---...((.(....).)).......))))).((((((.(((.((....)))))..))))))(((((((((.................))))))))). ( -20.65) >DroSim_CAF1 4992 113 - 1 AAUUUUAAAUAGGCUAAAA---AAUGGAUGAAAAACUUAUAAUAUAGGUACAUUUCAGUGCGCGAAAGCCACAAGAAAUUCAUCUAUUUAAAACAUAAUAAAAGAAAAGUGGGUGG ............(((...(---((((((((((..((((((...))))))...((((.(((.((....)))))..)))))))))))))))....(((............)))))).. ( -22.30) >DroYak_CAF1 10309 115 - 1 AGUCGUAUAUAAUUUAAAACUUAAUUGGUGAAGAACUUGUUAUAUAGCGAAAUUCCAUUGCGCGAAAGCCACAAGAAAUUAAUCCAUUUAAAACAUAAUAAAAGA-AAGUGGAAAG ..(((((((((((.....(((.....))).........))))))).))))..((((((((.((....)))........((((.....))))..............-.))))))).. ( -17.74) >consensus AAUUUUAAAUAAGCUAAAA___AAUGGAUGAAAAACUUAUAAUAUAGCUAAAUUUCAGUGCGCGAAAGCCACAAGAAAUUCAUCUAUUUAAAACAUAAUAAAAGAAAAGUGGAUGG ......................((((((((((...((........))..........(((.((....)))))......))))))))))............................ (-11.90 = -12.40 + 0.50)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:42 2006