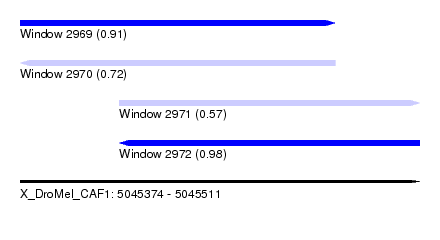
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,045,374 – 5,045,511 |
| Length | 137 |
| Max. P | 0.976913 |

| Location | 5,045,374 – 5,045,482 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.80 |
| Mean single sequence MFE | -14.37 |
| Consensus MFE | -12.40 |
| Energy contribution | -12.73 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5045374 108 + 22224390 CAAAAAAAAAGAAGA------GCCAAAAGCGCAGAAGAAAAUUUCGGUCUUAUUGCCAGUGUAAAAUUGUAAUUUUUACACCUUCACACCGCCCAAUAAACAACAAAAAAAAAG ..........((((.------((.....))((((((((.........)))).))))..((((((((.......))))))))))))............................. ( -15.10) >DroEre_CAF1 9388 94 + 1 CAA-----AAGAAGA------ACCAAAAACGUAGAAGGAAAUUUCGGUCUUAUUGCCAGUGUAA-AUUGUAAUUUUUACACCUUCACACCGCCCUAUAAACAAAAA-------- ...-----.......------.........((.(((((.......(((......)))..(((((-(........)))))))))))))...................-------- ( -13.70) >DroYak_CAF1 9645 101 + 1 AAA-----AAAAAAAAAAAAGAUCAAAAACGUAGAAGGAAAUUUCGGUCUUAUUGCCAGUGUAAAAUUGUAAUUUUUACACCUUCACACCGCCCAAUAAACAAAAC-------- ...-----......................((.(((((.......(((......)))..(((((((.......))))))))))))))...................-------- ( -14.30) >consensus CAA_____AAGAAGA______ACCAAAAACGUAGAAGGAAAUUUCGGUCUUAUUGCCAGUGUAAAAUUGUAAUUUUUACACCUUCACACCGCCCAAUAAACAAAAA________ .................................((((........(((......))).((((((((.......))))))))))))............................. (-12.40 = -12.73 + 0.33)

| Location | 5,045,374 – 5,045,482 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.80 |
| Mean single sequence MFE | -20.84 |
| Consensus MFE | -16.85 |
| Energy contribution | -17.85 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5045374 108 - 22224390 CUUUUUUUUUGUUGUUUAUUGGGCGGUGUGAAGGUGUAAAAAUUACAAUUUUACACUGGCAAUAAGACCGAAAUUUUCUUCUGCGCUUUUGGC------UCUUCUUUUUUUUUG ......(((((...((((((.(.((((((((((.((((.....)))).)))))))))).)))))))..)))))...........((.....))------............... ( -21.60) >DroEre_CAF1 9388 94 - 1 --------UUUUUGUUUAUAGGGCGGUGUGAAGGUGUAAAAAUUACAAU-UUACACUGGCAAUAAGACCGAAAUUUCCUUCUACGUUUUUGGU------UCUUCUU-----UUG --------.............(.(((((((((..((((.....)))).)-)))))))).)...((((((((((.............)))))).------))))...-----... ( -21.12) >DroYak_CAF1 9645 101 - 1 --------GUUUUGUUUAUUGGGCGGUGUGAAGGUGUAAAAAUUACAAUUUUACACUGGCAAUAAGACCGAAAUUUCCUUCUACGUUUUUGAUCUUUUUUUUUUUU-----UUU --------((((((((((((.(.((((((((((.((((.....)))).)))))))))).)))))))..))))))................................-----... ( -19.80) >consensus ________UUUUUGUUUAUUGGGCGGUGUGAAGGUGUAAAAAUUACAAUUUUACACUGGCAAUAAGACCGAAAUUUCCUUCUACGUUUUUGGU______UCUUCUU_____UUG ..............((((((.(.((((((((((.((((.....)))).)))))))))).))))))).((((((.............))))))...................... (-16.85 = -17.85 + 1.00)



| Location | 5,045,408 – 5,045,511 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 87.67 |
| Mean single sequence MFE | -18.21 |
| Consensus MFE | -11.64 |
| Energy contribution | -12.30 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5045408 103 + 22224390 AUUUCGGUCUUAUUGCCAGUGUAAAAUUGUAAUUUUUACACCUUCACACCGCCCAAUAAACAACAAAAAAAAAGGAAGAAAAAAAAGUGGCAUUUCCACUUUU .(((((((......))).((((((((.......))))))))....................................)))).((((((((.....)))))))) ( -18.30) >DroEre_CAF1 9417 91 + 1 AUUUCGGUCUUAUUGCCAGUGUAA-AUUGUAAUUUUUACACCUUCACACCGCCCUAUAAACAAAAA----------GGAA-AAAAAAUGGCAUUUCCACUUUU .....((......(((((((((((-(........)))))))...........(((..........)----------))..-......)))))...))...... ( -15.90) >DroYak_CAF1 9680 94 + 1 AUUUCGGUCUUAUUGCCAGUGUAAAAUUGUAAUUUUUACACCUUCACACCGCCCAAUAAACAAAAC--------GCGGAA-CAAAAAUGGCAUUUCCACUUUU .....((......(((((((((((((.......)))))))).......((((..............--------))))..-......)))))...))...... ( -20.44) >consensus AUUUCGGUCUUAUUGCCAGUGUAAAAUUGUAAUUUUUACACCUUCACACCGCCCAAUAAACAAAAA________G_GGAA_AAAAAAUGGCAUUUCCACUUUU .....((......(((((((((((((.......)))))))).......((..........................)).........)))))...))...... (-11.64 = -12.30 + 0.67)

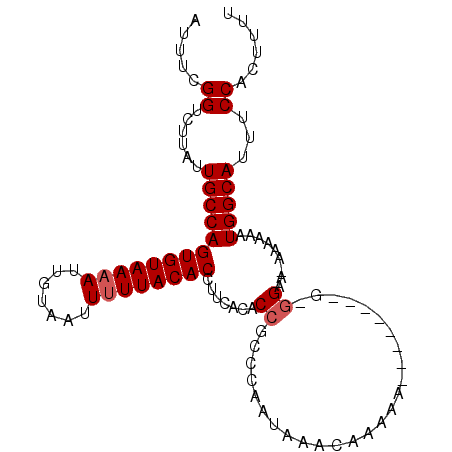
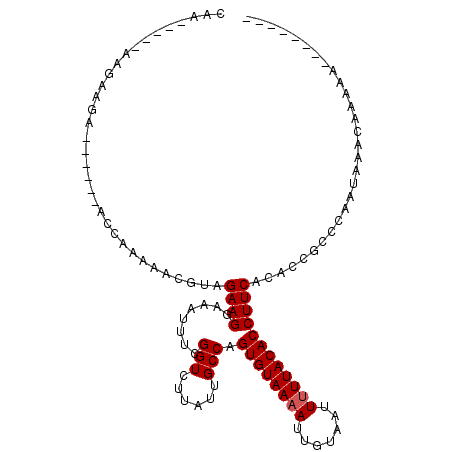
| Location | 5,045,408 – 5,045,511 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 87.67 |
| Mean single sequence MFE | -26.43 |
| Consensus MFE | -23.25 |
| Energy contribution | -23.37 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5045408 103 - 22224390 AAAAGUGGAAAUGCCACUUUUUUUUCUUCCUUUUUUUUUGUUGUUUAUUGGGCGGUGUGAAGGUGUAAAAAUUACAAUUUUACACUGGCAAUAAGACCGAAAU ((((((((.....))))))))..............(((((...((((((.(.((((((((((.((((.....)))).)))))))))).)))))))..))))). ( -28.20) >DroEre_CAF1 9417 91 - 1 AAAAGUGGAAAUGCCAUUUUUU-UUCC----------UUUUUGUUUAUAGGGCGGUGUGAAGGUGUAAAAAUUACAAU-UUACACUGGCAAUAAGACCGAAAU ((((((((.....)))))))).-....----------((((.((((....(.(((((((((..((((.....)))).)-)))))))).)....)))).)))). ( -23.50) >DroYak_CAF1 9680 94 - 1 AAAAGUGGAAAUGCCAUUUUUG-UUCCGC--------GUUUUGUUUAUUGGGCGGUGUGAAGGUGUAAAAAUUACAAUUUUACACUGGCAAUAAGACCGAAAU ((((((((.....)))))))).-...((.--------((((((((.....(.((((((((((.((((.....)))).)))))))))).))))))))))).... ( -27.60) >consensus AAAAGUGGAAAUGCCAUUUUUU_UUCC_C________UUUUUGUUUAUUGGGCGGUGUGAAGGUGUAAAAAUUACAAUUUUACACUGGCAAUAAGACCGAAAU ((((((((.....)))))))).....................((((....(.((((((((((.((((.....)))).)))))))))).)....))))...... (-23.25 = -23.37 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:40 2006