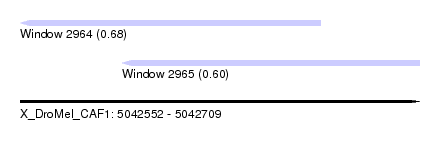
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,042,552 – 5,042,709 |
| Length | 157 |
| Max. P | 0.681808 |

| Location | 5,042,552 – 5,042,670 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.06 |
| Mean single sequence MFE | -18.96 |
| Consensus MFE | -14.44 |
| Energy contribution | -14.28 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
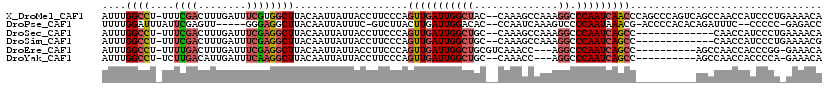
>X_DroMel_CAF1 5042552 118 - 22224390 UUACCUUCCCAGUUGAUUGGCUAC--CAAAGCCAAAGGCCCAAUCAACCCAGCCCAGUCAGCCAACCAUCCCUGAAAACAUCGAACCAGCCAAAGGAACAACAUCCACGAUCCACCAUUA ...((((....(((((((((....--....(((...)))))))))))).........((((..........)))).................))))........................ ( -16.80) >DroSec_CAF1 1448 105 - 1 UUACCUUCCCAGUUGAUUGGCUGC--CAAAGCCAAAGGCCCAAUCAGCC-------------CAACCAUCCCUGAAAACAUCGAACCAGCCAAAGGAGCAACAUCCACGAUCCACCAUUA ...........(((((((((..((--(.........)))))))))))).-------------................................(((......))).............. ( -18.30) >DroSim_CAF1 1527 105 - 1 UUACCUUCCCAGUUGAUUGGCUGC--CAAAGCCAAAGGCCCAAUCAGCC-------------CAACCAUCCCUGAAAACGUCGAACCAGCCAAAGGAGCAACAUCCACGAUCCACCAUUA ...........(((((((((..((--(.........)))))))))))).-------------................................(((......))).............. ( -18.30) >DroEre_CAF1 6617 103 - 1 UUACCUUCCCAGUUGAUUGGCUGCGUCAAACC---AGGCCCAAUCAGCC----------AGCCAACCACCCGG-GAAACAUCGCACCAGCCAAAGGA---ACAUCCACGAUCCACCAUUA .....(((((.((((..((((((.(((.....---.))).....)))))----------)..)))).....))-))).....((....))....(((---.(......).)))....... ( -23.20) >DroYak_CAF1 6499 101 - 1 UUACCUUCCCAGUUGAUUGGCUGC--CAAACC---AGGCCCAAUCAGCC----------AGCCAACCACCCCA-GAAACAUCGAACCAACCAAAGGA---ACAUCCACCAUCCACCGUUA ...((((....(((((((((..((--(.....---.)))))))))))).----------..............-((....))..........)))).---.................... ( -18.20) >consensus UUACCUUCCCAGUUGAUUGGCUGC__CAAAGCCAAAGGCCCAAUCAGCC__________AGCCAACCAUCCCUGAAAACAUCGAACCAGCCAAAGGA_CAACAUCCACGAUCCACCAUUA ...........(((((((((((..............)).)))))))))..............................................(((......))).............. (-14.44 = -14.28 + -0.16)

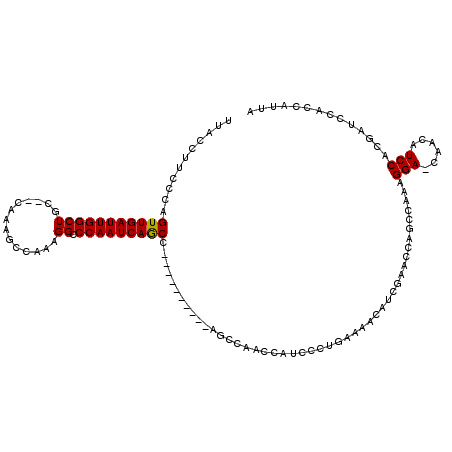
| Location | 5,042,592 – 5,042,709 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.43 |
| Mean single sequence MFE | -23.84 |
| Consensus MFE | -11.78 |
| Energy contribution | -12.39 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5042592 117 - 22224390 AUUUGGCCU-UUUCGACUUUGAUUUCGUGGCUUACAAUUAUUACCUUCCCAGUUGAUUGGCUAC--CAAAGCCAAAGGCCCAAUCAACCCAGCCCAGUCAGCCAACCAUCCCUGAAAACA ..(((((..-....((((((((((..(.(((((.(((((...........))))).((((((..--...))))))))))))))))))........)))).)))))............... ( -22.70) >DroPse_CAF1 1171 108 - 1 UUUUGGAUUUAUUCGAGUU-----GGGAGGCUUACAAUUAUUUC-GUCUUACUUGAUUGGACAC--CCAAUCAAAGUCCCCAAUAAACG-ACCCCACACAGAUUUC--CCCCC-GAGACC ((((((......(((.(((-----(((.(((((...........-(.....).(((((((....--))))))))))))))))))...))-).........(....)--...))-)))).. ( -23.30) >DroSec_CAF1 1488 104 - 1 AUUUGGCCU-UUUCGACUUUGAUUUCGAGGCUUACAAUUAUUACCUUCCCAGUUGAUUGGCUGC--CAAAGCCAAAGGCCCAAUCAGCC-------------CAACCAUCCCUGAAAACA ....(((((-((((((........))))(((...((((((.............))))))...))--)......)))))))...((((..-------------.........))))..... ( -22.72) >DroSim_CAF1 1567 104 - 1 AUUUGGCCU-UUUCGACUUUGAUUUCGAGGCUUACAAUUAUUACCUUCCCAGUUGAUUGGCUGC--CAAAGCCAAAGGCCCAAUCAGCC-------------CAACCAUCCCUGAAAACG ....(((((-((((((........))))(((...((((((.............))))))...))--)......)))))))...((((..-------------.........))))..... ( -22.72) >DroEre_CAF1 6654 105 - 1 AUUUGGCCU-UUUUGACUUUGAUUUCGAGGCUUACAAUUAUUACCUUCCCAGUUGAUUGGCUGCGUCAAACC---AGGCCCAAUCAGCC----------AGCCAACCACCCGG-GAAACA ....(((((-(...((........)))))))).............(((((.((((..((((((.(((.....---.))).....)))))----------)..)))).....))-)))... ( -25.50) >DroYak_CAF1 6536 103 - 1 AUUUGGCCU-UCUUGACAUUGAUUUCAAGGCUUACAAUUAUUACCUUCCCAGUUGAUUGGCUGC--CAAACC---AGGCCCAAUCAGCC----------AGCCAACCACCCCA-GAAACA ..(((((..-((((((........)))))).....................(((((((((..((--(.....---.)))))))))))).----------.)))))........-...... ( -26.10) >consensus AUUUGGCCU_UUUCGACUUUGAUUUCGAGGCUUACAAUUAUUACCUUCCCAGUUGAUUGGCUGC__CAAAGCCAAAGGCCCAAUCAGCC__________AGCCAACCACCCCU_AAAACA ....((((....((((........))))))))...................(((((((((((..............)).)))))))))................................ (-11.78 = -12.39 + 0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:34 2006