
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,042,126 – 5,042,225 |
| Length | 99 |
| Max. P | 0.500000 |

| Location | 5,042,126 – 5,042,225 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.33 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -23.54 |
| Energy contribution | -23.73 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
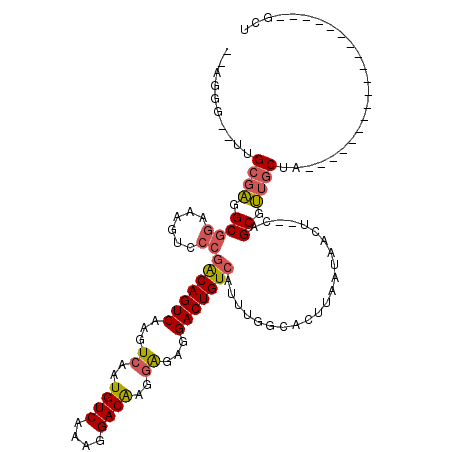
>X_DroMel_CAF1 5042126 99 + 22224390 GGCGGUUGUUGCGAGGCGGAAAGUCCCGACAGUCAAGUCAAUGUCAAAGGACAAGGAGAAGACUGUCAUUUGGCACUUAAUAACU--CAGCGUUGCUA-------------------GCU ((((((...((((((..(..((((((.(((((((...((..((((....))))..))...)))))))....)).))))..)..))--).)))..))).-------------------))) ( -29.60) >DroPse_CAF1 726 111 + 1 --GCCC--CCGGCGGGCGGAAG-----AGCAGUCAAGUCAAUGUCAAAGGACGAACGGAGGACUGUCAUUUGGCACUUAAUAACUCUCAGCGCUGCUCCCAACCCCACCCCCCCGAUGCU --((..--(.((.(((.((..(-----((((((.........(((....)))(...((((...((((....))))........))))...))))))))......)).))).)).)..)). ( -28.50) >DroSec_CAF1 1035 97 + 1 --UGGGUCUUGCGAGGCGGAAAGUCCCGGCAGUCAAGUCAAUGUCAAAGGACAAGGAGGGGACUGUCAUUUGGCACUUAAUAACU--CAGCGUUGCUA-------------------GCU --..(((..((((((..(..((((((.(((((((...((..((((....))))..))...)))))))....)).))))..)..))--).)))..))).-------------------... ( -29.00) >DroSim_CAF1 1112 97 + 1 --CGGGUCUUGCGAGGCGGAAAGUCCCGGCAGUCAAGUCAAUGUCAAAGGACAAGGAGGGGACUGUCAUUUGGCACUUAAUAACU--CAGCGUUGCUA-------------------GCU --..(((..((((((..(..((((((.(((((((...((..((((....))))..))...)))))))....)).))))..)..))--).)))..))).-------------------... ( -29.00) >DroEre_CAF1 6204 95 + 1 --AGGG--CUGCGAGGCGGAAAGUCCCGACAGUCAAGUCAAUGUCAAAGGACAAGGAGAGGACUGUCAUUUGGCACUUAAUAACU--CAGCGUUGCUA-------------------GCU --..((--((((((.((((......))(((((((...((..((((....))))..))...)))))))..................--..)).)))).)-------------------))) ( -32.70) >DroYak_CAF1 6098 95 + 1 --AGGG--UUGCGAUGCGGAAAGUCCCGACAGUCAAGUCUAUGUCAAAGGACAAGGAGAGGACUGUCAUUUGGCACUUAAUAACU--CAGCGUUGCUA-------------------GCU --..((--(((((((((((......))(((((((...(((.((((....)))).)))...)))))))..................--..))))))).)-------------------))) ( -33.50) >consensus __AGGG__UUGCGAGGCGGAAAGUCCCGACAGUCAAGUCAAUGUCAAAGGACAAGGAGAGGACUGUCAUUUGGCACUUAAUAACU__CAGCGUUGCUA___________________GCU ..........((((.((((......))(((((((...((..((((....))))..))...)))))))......................)).))))........................ (-23.54 = -23.73 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:32 2006