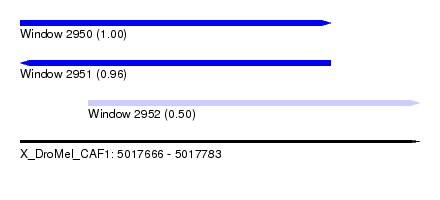
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,017,666 – 5,017,783 |
| Length | 117 |
| Max. P | 0.996850 |

| Location | 5,017,666 – 5,017,757 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.21 |
| Mean single sequence MFE | -32.03 |
| Consensus MFE | -18.02 |
| Energy contribution | -18.85 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5017666 91 + 22224390 -----------UAUGGGCCAUGUAAAGUUGAACGUGGGCAUAACAAUGUGCCCUUAACUUGGCCCACAGACUGCACUUUUGUGCCUCGUCUCUGUCUUACCA------ -----------..(((((((.....((((((....(((((((....)))))))))))))))))))).((((.((((....))))...))))...........------ ( -33.40) >DroPse_CAF1 112375 94 + 1 GAAGGCUGGGACGAGUUCCAUGUAAACUUUAACGUGGGCAUAACAAUGUGCCCUUAACUUGGCCCAGACACUGCU-------GCUGCAGUUCUUUCCUCCC------- ((((((((((.((((((..((((........))))(((((((....)))))))..)))))).)))))..(((((.-------...))))).))))).....------- ( -34.40) >DroSec_CAF1 83597 97 + 1 -----------UAUGGGCCAUGUAAAGUUGAACGUGGGCAUAACAAUGUGCCCUUAACUUGGCCCACAGACUGCACUUUUGUGCCUCGUCUCUGUCUUACCCUUGUAC -----------..(((((((.....((((((....(((((((....)))))))))))))))))))).((((.((((....))))...))))................. ( -33.40) >DroEre_CAF1 87997 96 + 1 -----------UAUGGGCCAUGUAAAGUUGAACGUGGGCAUAACAAUGUGCCGUUAACUUGGCCCAUAGACUGCAGUUUUGUACCUCGUCUCUGUCUUACCCUU-CAC -----------(((((((((.....((((((.....((((((....)))))).)))))))))))))))....((((...((.....))...)))).........-... ( -27.50) >DroYak_CAF1 85062 96 + 1 -----------UACGGGCCAUGUAAAGUUGAACGUGGGCAUAACAAUGUGCCCUUAACUUGGCCCAUAGACUGCAGUUUUGUGCCCCGUCUCUGUCUUACCCAU-CAC -----------...((((((.....((((((....(((((((....)))))))))))))))))))..((((.(((......)))...)))).............-... ( -29.10) >DroPer_CAF1 104062 94 + 1 GAAGGCUGGGACGAGUUCCAUGUAAACUUUAACGUGGGCAUAACAAUGUGCCCUUAACUUGGCCCAGACACUGCU-------GCUGCAGUUCUUUCCUCCC------- ((((((((((.((((((..((((........))))(((((((....)))))))..)))))).)))))..(((((.-------...))))).))))).....------- ( -34.40) >consensus ___________UAUGGGCCAUGUAAAGUUGAACGUGGGCAUAACAAUGUGCCCUUAACUUGGCCCACAGACUGCA_UUUUGUGCCUCGUCUCUGUCUUACCC______ ..............(((((((((........))))(((((((....))))))).......)))))........................................... (-18.02 = -18.85 + 0.83)

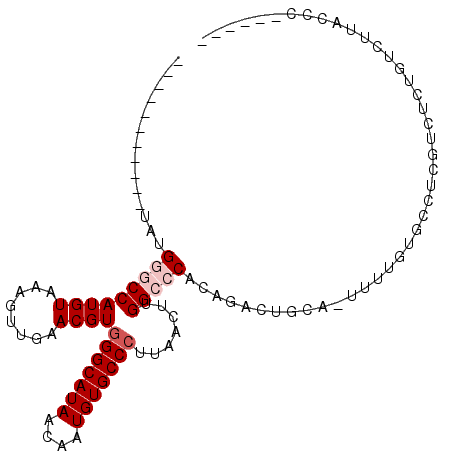
| Location | 5,017,666 – 5,017,757 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 75.21 |
| Mean single sequence MFE | -30.43 |
| Consensus MFE | -18.74 |
| Energy contribution | -20.68 |
| Covariance contribution | 1.95 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5017666 91 - 22224390 ------UGGUAAGACAGAGACGAGGCACAAAAGUGCAGUCUGUGGGCCAAGUUAAGGGCACAUUGUUAUGCCCACGUUCAACUUUACAUGGCCCAUA----------- ------...........((((...((((....)))).))))((((((((......(((((........)))))..((........)).)))))))).----------- ( -33.60) >DroPse_CAF1 112375 94 - 1 -------GGGAGGAAAGAACUGCAGC-------AGCAGUGUCUGGGCCAAGUUAAGGGCACAUUGUUAUGCCCACGUUAAAGUUUACAUGGAACUCGUCCCAGCCUUC -------((((.((.((((((((...-------.))))).)))(..(((......(((((........)))))..((........)).)))..))).))))....... ( -30.10) >DroSec_CAF1 83597 97 - 1 GUACAAGGGUAAGACAGAGACGAGGCACAAAAGUGCAGUCUGUGGGCCAAGUUAAGGGCACAUUGUUAUGCCCACGUUCAACUUUACAUGGCCCAUA----------- .................((((...((((....)))).))))((((((((......(((((........)))))..((........)).)))))))).----------- ( -33.60) >DroEre_CAF1 87997 96 - 1 GUG-AAGGGUAAGACAGAGACGAGGUACAAAACUGCAGUCUAUGGGCCAAGUUAACGGCACAUUGUUAUGCCCACGUUCAACUUUACAUGGCCCAUA----------- ...-.............((((..(((.....)))...))))(((((((((((((((((((........))))...))).)))).....)))))))).----------- ( -25.60) >DroYak_CAF1 85062 96 - 1 GUG-AUGGGUAAGACAGAGACGGGGCACAAAACUGCAGUCUAUGGGCCAAGUUAAGGGCACAUUGUUAUGCCCACGUUCAACUUUACAUGGCCCGUA----------- ...-.............((((...(((......))).))))((((((((......(((((........)))))..((........)).)))))))).----------- ( -29.60) >DroPer_CAF1 104062 94 - 1 -------GGGAGGAAAGAACUGCAGC-------AGCAGUGUCUGGGCCAAGUUAAGGGCACAUUGUUAUGCCCACGUUAAAGUUUACAUGGAACUCGUCCCAGCCUUC -------((((.((.((((((((...-------.))))).)))(..(((......(((((........)))))..((........)).)))..))).))))....... ( -30.10) >consensus ______GGGUAAGACAGAGACGAGGCACAAAA_UGCAGUCUAUGGGCCAAGUUAAGGGCACAUUGUUAUGCCCACGUUCAACUUUACAUGGCCCAUA___________ .................((((...(((......))).))))((((((((......(((((........)))))..((........)).))))))))............ (-18.74 = -20.68 + 1.95)



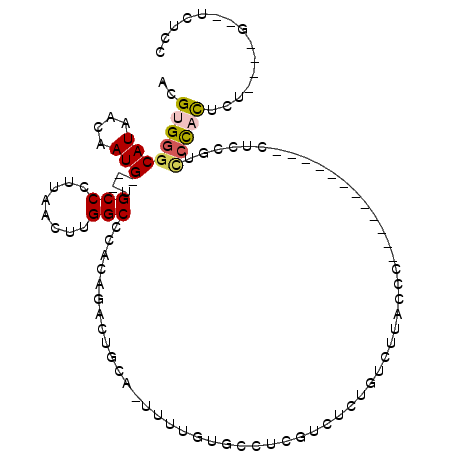
| Location | 5,017,686 – 5,017,783 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 65.67 |
| Mean single sequence MFE | -20.87 |
| Consensus MFE | -4.91 |
| Energy contribution | -5.13 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.24 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 5017686 97 + 22224390 ACGUGGGCAUAACAAUG---UGCCCUUAACUUGGCCCACAGACUGCACUUUUGUGCCUCGUCUCUGUCUUACCA------------CUCCGUCCCACUCUUUACGACUCUCU .((((((((((....))---)))))......(((..((.((((.((((....))))...)))).)).....)))------------................)))....... ( -22.80) >DroPse_CAF1 112406 79 + 1 ACGUGGGCAUAACAAUG---UGCCCUUAACUUGGCCCAGACACUGCU-------GCUGCAGUUCUUUCCUCCC-------------CUCUCUCCCCC----------UCUCC ....(((((((....))---))))).......((...(((.(((((.-------...))))))))..))....-------------...........----------..... ( -19.70) >DroSec_CAF1 83617 109 + 1 ACGUGGGCAUAACAAUG---UGCCCUUAACUUGGCCCACAGACUGCACUUUUGUGCCUCGUCUCUGUCUUACCCUUGUACAGCCGUCUCCGUCCCACUCUUCUCGACUCUCU .((.(((((((....))---)))))......(((..(..((((.((((....)))).......(((((........).))))..))))..)..))).......))....... ( -24.10) >DroEre_CAF1 88017 108 + 1 ACGUGGGCAUAACAAUG---UGCCGUUAACUUGGCCCAUAGACUGCAGUUUUGUACCUCGUCUCUGUCUUACCCUU-CACAGCCGUCUGCGUUCCACUCUCCCAGACUCUUU .....(((......(((---.((((......)))).))).....((((...((.....))...)))).........-....)))(((((.(.........).)))))..... ( -20.10) >DroWil_CAF1 88743 97 + 1 ACGUGAGCAUAACAAUGUUGUGCCCUUAAGUUGGCCCU--GGCUGCAGCUUCGCCAUUCCUUUCUACCUUCACCUU-------CGUC----UUCUACUUCCUCUG--UCUUC ..(((((((((((...)))))))........((((...--(((....)))..)))).............))))...-------....----..............--..... ( -18.40) >DroPer_CAF1 104093 79 + 1 ACGUGGGCAUAACAAUG---UGCCCUUAACUUGGCCCAGACACUGCU-------GCUGCAGUUCUUUCCUCCC-------------CUCUCUGCCCU----------UCUCC ....(((((......((---.(((........))).))((.(((((.-------...))))))).........-------------.....))))).----------..... ( -20.10) >consensus ACGUGGGCAUAACAAUG___UGCCCUUAACUUGGCCCACAGACUGCA_UUUUGUGCCUCGUCUCUGUCUUACCC____________CUCCGUCCCACUCU____G__UCUCC ..((((((((....)))....(((........))).........................................................)))))............... ( -4.91 = -5.13 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:21 2006