
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 5,016,099 – 5,016,201 |
| Length | 102 |
| Max. P | 0.976465 |

| Location | 5,016,099 – 5,016,201 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 81.15 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -16.84 |
| Energy contribution | -17.84 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
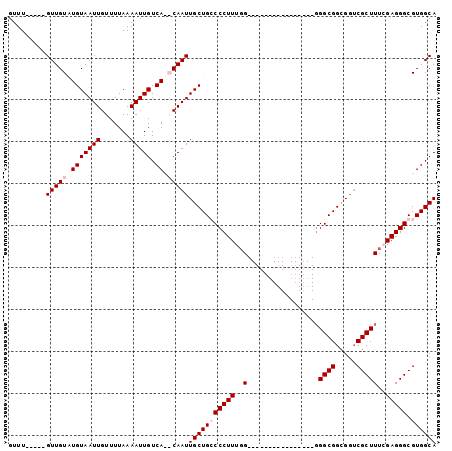
>X_DroMel_CAF1 5016099 102 + 22224390 GUUUGUUUUGUUGUAUGUAAUUGUUUUAAAAUUGUCACACAAUUGCUGCCCCUUUUGGUGGGGGGGGGGUGGUGGGCGGCGGUCGCUUUCGAGGGCGUGGCA ..........((((.(((((((.......))))).)).))))(..(..((((((((.....))))))))..)..)((.(((.((........)).))).)). ( -31.20) >DroSec_CAF1 82174 86 + 1 GUUUGUUUUGUUGUAUGUAAUUGUUUUAAAAUUGUCACACAAUUGCUGCCCCUUUGG----------------GGGCGGCGGUCGCUUUCGAGGGCGUGGCA ................................((((((...(((((((((((....)----------------)))))))))).((((....)))))))))) ( -30.10) >DroEre_CAF1 86481 79 + 1 GUUU-----GUUGUGUGUAAUUGUUUUAAAAUUGUCA--CAAUUGCUGGCCCUUUGG----------------GGGCGGUGGUCGCUUUGGAGGGCGUGGCA ....-----....(((((((((.......))))).))--))..(((((((((((..(----------------((.((.....)))))..)))))).))))) ( -26.80) >DroYak_CAF1 83703 78 + 1 GUUU-----GUUGUUUGUAAUUGUUUUAAAAUUGUCA--CAAUUGCUGGCCCUUUGG-----------------GGCGGUGUACGCUUUGGAGGGCGUGGCA ...(-----((..(..((((((((..((....))..)--)))))))..((((((..(-----------------((((.....)))))..)))))))..))) ( -31.20) >consensus GUUU_____GUUGUAUGUAAUUGUUUUAAAAUUGUCA__CAAUUGCUGCCCCUUUGG________________GGGCGGCGGUCGCUUUCGAGGGCGUGGCA .........(((((.(((((((.......))))).)).)))))(((((((((((..(.................((((.....)))))..))))).)))))) (-16.84 = -17.84 + 1.00)


| Location | 5,016,099 – 5,016,201 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 81.15 |
| Mean single sequence MFE | -17.01 |
| Consensus MFE | -8.06 |
| Energy contribution | -9.55 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
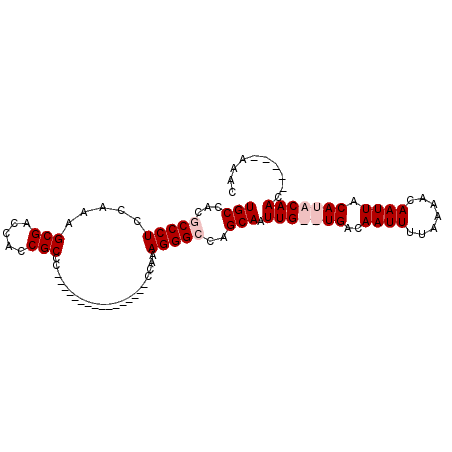
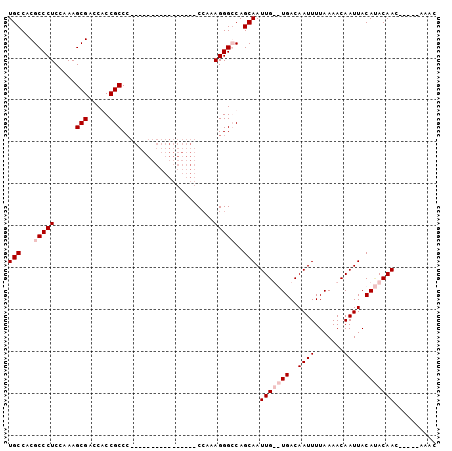
>X_DroMel_CAF1 5016099 102 - 22224390 UGCCACGCCCUCGAAAGCGACCGCCGCCCACCACCCCCCCCCCACCAAAAGGGGCAGCAAUUGUGUGACAAUUUUAAAACAAUUACAUACAACAAAACAAAC ((.(((((.(.(....).)...((.((((......................)))).))....))))).))................................ ( -16.45) >DroSec_CAF1 82174 86 - 1 UGCCACGCCCUCGAAAGCGACCGCCGCCC----------------CCAAAGGGGCAGCAAUUGUGUGACAAUUUUAAAACAAUUACAUACAACAAAACAAAC ((.(((((.(.(....).)...((.((((----------------(....))))).))....))))).))................................ ( -21.10) >DroEre_CAF1 86481 79 - 1 UGCCACGCCCUCCAAAGCGACCACCGCCC----------------CCAAAGGGCCAGCAAUUG--UGACAAUUUUAAAACAAUUACACACAAC-----AAAC .((...(((((.....(((.....)))..----------------....)))))..))(((((--(............)))))).........-----.... ( -15.20) >DroYak_CAF1 83703 78 - 1 UGCCACGCCCUCCAAAGCGUACACCGCC-----------------CCAAAGGGCCAGCAAUUG--UGACAAUUUUAAAACAAUUACAAACAAC-----AAAC .((...(((((.....(((.....))).-----------------....)))))..))(((((--(............)))))).........-----.... ( -15.30) >consensus UGCCACGCCCUCCAAAGCGACCACCGCCC________________CCAAAGGGCCAGCAAUUG__UGACAAUUUUAAAACAAUUACAUACAAC_____AAAC (((...(((((.....(((.....)))......................)))))..))).(((((((..((((.......)))).))))))).......... ( -8.06 = -9.55 + 1.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:18 2006