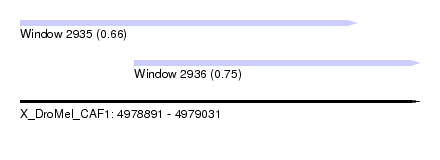
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,978,891 – 4,979,031 |
| Length | 140 |
| Max. P | 0.754188 |

| Location | 4,978,891 – 4,979,009 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.43 |
| Mean single sequence MFE | -24.15 |
| Consensus MFE | -14.95 |
| Energy contribution | -15.20 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
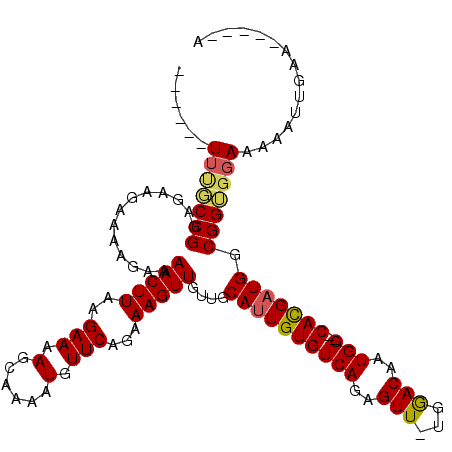
>X_DroMel_CAF1 4978891 118 + 22224390 UUUUUGUUUGCCGUUGAAGAAAAGAAAACUUAAGAAAAGCAAAAUGUUCAGAAAGUUGUUGCAUUGUGUCAGAGUU-UGGACAAUGGUGCACGAUGGCGGUGGAAAAAUUGAACCAAAA (((((..(((((((((..........(((((......(((.....)))....)))))..(((((..((((......-..))).)..))))))))))))))..)))))............ ( -24.80) >DroSec_CAF1 44421 107 + 1 ------UUUGCCGUGGAAGAUAGGAAAACUUAAGAAAAGCAAAAUGUUCAGAAAGUUGUUGCAUUGUGUCAGAGUU-UGAACAAUGGUGCACGAUGGCGGUGGAAAAAUUGAA-----A ------((..((((.(..(.((((....))))......(((...((((((((...(((.(((...))).)))..))-))))))....))).)..).))))..)).........-----. ( -22.80) >DroEre_CAF1 49906 105 + 1 ------UACCCCGGAGAAAAAAAGAAAACUUAGGAAAAGCAAAAUGUUCAGAAAGUUGUUGCAUUGUGUCAGAGUU-UGGACAAUGG--CACGAUGGCGGGGGAAAAAUUGAA-----A ------..(((((.............(((((..(((.(......).)))...)))))....((((((((((..((.-...))..)))--))))))).)))))...........-----. ( -27.10) >DroYak_CAF1 46618 104 + 1 ------UU--CCGGCAUAGAAAAGAAAACUUAAGAAAAGCUAAAUUUUCGGAAAGUUGUUGCAUUGUGUCAGAGUUUUGGACAAUGG--CAUGUUGGCGGUGGAAAAAUUGAU-----U ------((--(((.(...........(((((..(((((......)))))...)))))...((..(((((((..(((...)))..)))--))))...))).)))))........-----. ( -21.90) >consensus ______UUUGCCGGAGAAGAAAAGAAAACUUAAGAAAAGCAAAAUGUUCAGAAAGUUGUUGCAUUGUGUCAGAGUU_UGGACAAUGG__CACGAUGGCGGUGGAAAAAUUGAA_____A ......(((((((.............(((((..(((.(......).)))...)))))....((((((((((..(((...)))..)))..))))))).)))))))............... (-14.95 = -15.20 + 0.25)

| Location | 4,978,931 – 4,979,031 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 84.75 |
| Mean single sequence MFE | -16.97 |
| Consensus MFE | -12.47 |
| Energy contribution | -12.47 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
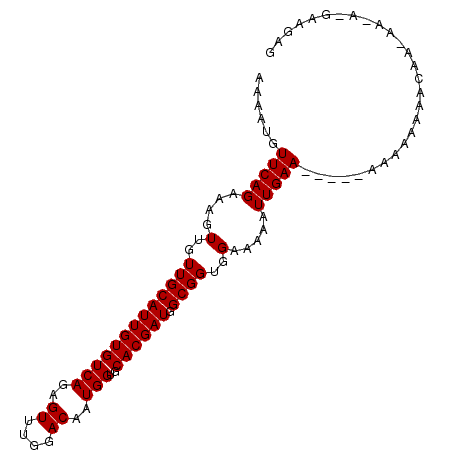
>X_DroMel_CAF1 4978931 100 + 22224390 AAAAUGUUCAGAAAGUUGUUGCAUUGUGUCAGAGUUUGGACAAUGGUGCACGAUGGCGGUGGAAAAAUUGAACCAAAAAAAAAAAAAAGAAAAGGAAGAG .....((((((....(..(((((((((((((..((....))..))..))))))).))))..).....))))))........................... ( -17.60) >DroSec_CAF1 44455 95 + 1 AAAAUGUUCAGAAAGUUGUUGCAUUGUGUCAGAGUUUGAACAAUGGUGCACGAUGGCGGUGGAAAAAUUGAA-----AAAAAAAACAAAAAUACGUAGAG ......(((((....(..((((((((((.((..(((.....)))..)))))))).))))..).....)))))-----....................... ( -15.00) >DroEre_CAF1 49940 84 + 1 AAAAUGUUCAGAAAGUUGUUGCAUUGUGUCAGAGUUUGGACAAUGG--CACGAUGGCGGGGGAAAAAUUGAA-----AAAAAAACC---------AAGAG ......(((((....((.(((((((((((((..((....))..)))--)))))).)))).)).....)))))-----.........---------..... ( -18.30) >consensus AAAAUGUUCAGAAAGUUGUUGCAUUGUGUCAGAGUUUGGACAAUGGUGCACGAUGGCGGUGGAAAAAUUGAA_____AAAAAAAACAA_AA_A_GAAGAG ......(((((....(..(((((((((((((..((....))..)))..)))))).))))..).....)))))............................ (-12.47 = -12.47 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:19:07 2006