
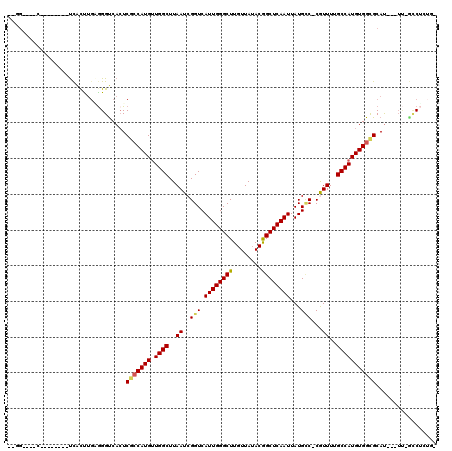
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,937,936 – 4,938,063 |
| Length | 127 |
| Max. P | 0.881993 |

| Location | 4,937,936 – 4,938,037 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.78 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -21.76 |
| Energy contribution | -21.90 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
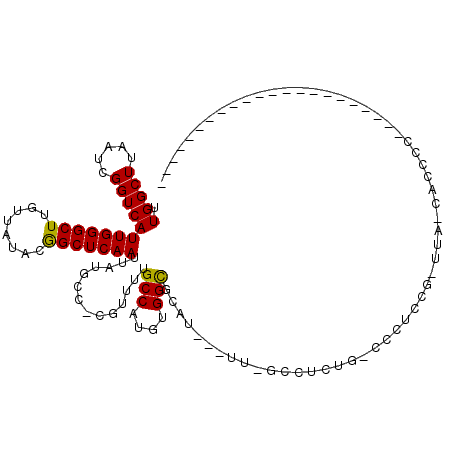
>X_DroMel_CAF1 4937936 101 + 22224390 --GG----C--------UCACUUGAGGGUCACUCGCCAUGUUGGCUUAAUCGGUCAUUGGGCUUGUUAUACGGCUCAAUUAUGCC-CGUUUUGCCAUGUGGCGCAU---UU-GCCUCUGG --((----(--------......(((.....)))(((((((.(((..(((.(((.((((((((........))))))))...)))-.)))..))))))))))....---..-)))..... ( -36.10) >DroPse_CAF1 11789 114 + 1 --UGGGGGCCAAAACAUCCGUCCGUGGGUCAGUCGACAUGCUGGCUUAAUCGGUCAUUGGGCUUGUUAGACGGCUCAAUUAUGCCCCGUUUUGCCAUGUGGCGCAU---UU-CUCUCUCC --..(((((..........(.((((((((((((......)))))))))..))).)((((((((........))))))))...))))).....(((....)))....---..-........ ( -37.40) >DroEre_CAF1 12328 96 + 1 --GG----C--------UCACUUGAGGGUCACUCGCCAUGUUGGCUUAAUCGGUCAUUGGGCUUGUUAUACGGCUCAAUUAUGCC-CGUUUUGCCAUGUGGCGCAU---UU-GCC----- --((----(--------((......)))))...((((((((.(((..(((.(((.((((((((........))))))))...)))-.)))..)))))))))))...---..-...----- ( -34.90) >DroWil_CAF1 11544 104 + 1 --UU----G--------UUGUUUGGGGGUCACUCUCCAUGUUGGCUUAAUCGGUCAUUGGGCUUGUUAUACAGCUCAAUUAUGUC-CAUUUUGCCAUGUGGCGCACCCUUG-AGCUCUGC --..----.--------..(((..(((((....(.((((((.(((..(((.((.(((((((((.(.....)))))))...))).)-))))..))))))))).).)))))..-)))..... ( -35.80) >DroYak_CAF1 11746 101 + 1 --GG----C--------UCACUUGAGGGUCACUCGCCAUGUUGGCUUAAUCGGUCAUUGGGCUUGUUAUACGGCUCAAUUAUGCC-CGUUUUGCCAUGUGGCGCAU---UU-GCCUGUGG --((----(--------......(((.....)))(((((((.(((..(((.(((.((((((((........))))))))...)))-.)))..))))))))))....---..-)))..... ( -36.10) >DroAna_CAF1 11723 101 + 1 CAAG----U--------UCACUUUUAGGCCACUCGCCAUGUUGGCUUAAUCGGUCAUUGGGCUUGUUAUACGGCUCAAUUAUGGC-CAUUUGGCCAUGUGGUGCAU---GCAGCCAC--- ....----.--------.........(((((..((((((((.((((.(((.((((((((((((........))))))...)))))-)))).))))))))))))..)---)..)))..--- ( -35.70) >consensus __GG____C________UCACUUGAGGGUCACUCGCCAUGUUGGCUUAAUCGGUCAUUGGGCUUGUUAUACGGCUCAAUUAUGCC_CGUUUUGCCAUGUGGCGCAU___UU_GCCUCUG_ .................................(((((((.((((..((..(((.((((((((........))))))))...)))...))..)))))))))))................. (-21.76 = -21.90 + 0.14)


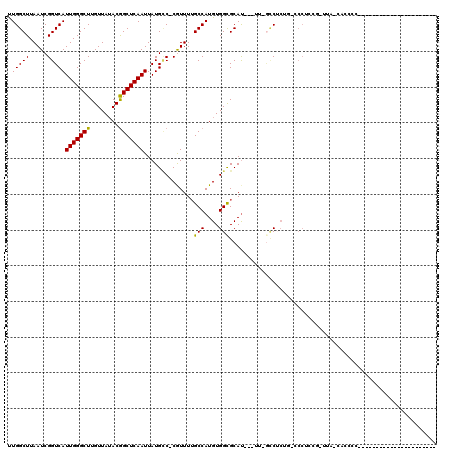
| Location | 4,937,962 – 4,938,063 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.85 |
| Mean single sequence MFE | -25.18 |
| Consensus MFE | -14.53 |
| Energy contribution | -14.25 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.680600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4937962 101 + 22224390 UUGGCUUAAUCGGUCAUUGGGCUUGUUAUACGGCUCAAUUAUGCC-CGUUUUGCCAUGUGGCGCAU---UU-GCCUCUGGCCCUCUGGUUA-CACCCCCCUCCCCAC------------- .((((..(((.(((.((((((((........))))))))...)))-.)))..))))((((((.((.---..-(((...)))....))))))-)).............------------- ( -27.20) >DroPse_CAF1 11827 93 + 1 CUGGCUUAAUCGGUCAUUGGGCUUGUUAGACGGCUCAAUUAUGCCCCGUUUUGCCAUGUGGCGCAU---UU-CUCUCUCCACCCCAC-UCACCCCUGG---------------------- .((((..(((.((.(((((((((........))))))...))).)).)))..)))).((((.(...---..-....).)))).(((.-.......)))---------------------- ( -20.30) >DroSim_CAF1 3213 91 + 1 UUGGCUUAAUCGGUCAUUGGGCUUGUUAUACGGCUCAAUUAUGCC-CGUUUUGCCAUGUGGCGCAU---UU-GCCUCUGGCCCUCUG-UUA-CAACCC---------------------- .((((..(((.(((.((((((((........))))))))...)))-.)))..))))(((((((...---..-(((...)))....))-)))-))....---------------------- ( -27.10) >DroEre_CAF1 12354 83 + 1 UUGGCUUAAUCGGUCAUUGGGCUUGUUAUACGGCUCAAUUAUGCC-CGUUUUGCCAUGUGGCGCAU---UU-GCC--------UCCG-GUC-CUCUGG---------------------- .((((..(((.(((.((((((((........))))))))...)))-.)))..))))...((((...---.)-)))--------.(((-(..-..))))---------------------- ( -26.10) >DroWil_CAF1 11570 110 + 1 UUGGCUUAAUCGGUCAUUGGGCUUGUUAUACAGCUCAAUUAUGUC-CAUUUUGCCAUGUGGCGCACCCUUG-AGCUCUGCUCCUCC--UUC-CAACUC-----CCAAAACUUCCUGCUUC .((((..(((.((.(((((((((.(.....)))))))...))).)-))))..)))).(..(.((.......-.)).)..)......--...-......-----................. ( -22.80) >DroAna_CAF1 11751 84 + 1 UUGGCUUAAUCGGUCAUUGGGCUUGUUAUACGGCUCAAUUAUGGC-CAUUUGGCCAUGUGGUGCAU---GCAGCCAC---CCC----------AC--C----CCCAC------------- .(((((.(((.((((((((((((........))))))...)))))-)))).))))).(((((....---...)))))---...----------..--.----.....------------- ( -27.60) >consensus UUGGCUUAAUCGGUCAUUGGGCUUGUUAUACGGCUCAAUUAUGCC_CGUUUUGCCAUGUGGCGCAU___UU_GCCUCUG_CCCUCCG_UUA_CACCCC______________________ .(((((.....)))))(((((((........)))))))..............(((....))).......................................................... (-14.53 = -14.25 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:56 2006