
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,937,156 – 4,937,250 |
| Length | 94 |
| Max. P | 0.834128 |

| Location | 4,937,156 – 4,937,250 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 85.19 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -19.36 |
| Energy contribution | -19.68 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4937156 94 + 22224390 GCCAAA--CCGGCAGAUAAAUUGUAGAUACAGACAGGUACACAGGUGCACACACCC--AUAUAGCGUAUAGCAAAAGG-----CGGAAUGUGUGCCAAGUGGG ......--((.((.......((((....))))...((((((((((((....)))).--.....((.....))......-----.....))))))))..)).)) ( -24.30) >DroSim_CAF1 2412 94 + 1 GCCAAA--CCGGCAGAUAAAUUGUAGAUACAGACAGGUACACAGGUGCACACACCU--AUAUAGCAUAUAGCAAAAGG-----CGGAAUGUGUGCCAAGUGGG ......--((.((.......((((....))))...((((((((((((....)))))--.....((.....))......-----......)))))))..)).)) ( -24.20) >DroEre_CAF1 11465 94 + 1 GCCAAA--CCGGCAGAUAAAUUGUAGAUACAGACAGGUACACAGGUGCACACAUCCACAUAUA-------GCAAUAGCGGUAUGGGAAUGUGUGCCAAGUGGG (((...--..))).......((((........))))...(((..(.((((((((((.(((((.-------((....)).))))))).)))))))))..))).. ( -30.80) >DroYak_CAF1 10928 94 + 1 GGCAAACACCGGCAGAUAAAUUGUAGAUACAGGCAGGUACACAGGUGCACACACCC--AUAUA-------GCAAAAGCGGAAUGGAAAUGUGUGCCAAGUGGG ........((.((.......((((....))))...((((((((((((....))))(--((...-------((....))...)))....))))))))..)).)) ( -24.90) >consensus GCCAAA__CCGGCAGAUAAAUUGUAGAUACAGACAGGUACACAGGUGCACACACCC__AUAUA_______GCAAAAGC_____CGGAAUGUGUGCCAAGUGGG ........((.((.......((((....))))...((((((((((((....))))...............((....))..........))))))))..)).)) (-19.36 = -19.68 + 0.31)

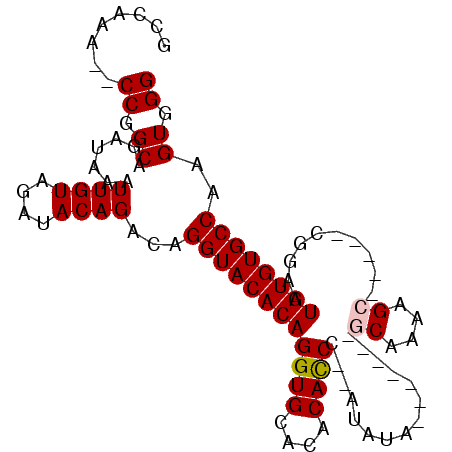
| Location | 4,937,156 – 4,937,250 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 85.19 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -15.90 |
| Energy contribution | -15.65 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4937156 94 - 22224390 CCCACUUGGCACACAUUCCG-----CCUUUUGCUAUACGCUAUAU--GGGUGUGUGCACCUGUGUACCUGUCUGUAUCUACAAUUUAUCUGCCGG--UUUGGC .(((.((((((........(-----(.....))((((.(((((((--(((((....))))))))))...)).)))).............))))))--..))). ( -20.80) >DroSim_CAF1 2412 94 - 1 CCCACUUGGCACACAUUCCG-----CCUUUUGCUAUAUGCUAUAU--AGGUGUGUGCACCUGUGUACCUGUCUGUAUCUACAAUUUAUCUGCCGG--UUUGGC .(((.((((((........(-----(.....))((((.(((((((--(((((....))))))))))...)).)))).............))))))--..))). ( -22.10) >DroEre_CAF1 11465 94 - 1 CCCACUUGGCACACAUUCCCAUACCGCUAUUGC-------UAUAUGUGGAUGUGUGCACCUGUGUACCUGUCUGUAUCUACAAUUUAUCUGCCGG--UUUGGC ..(((...((((((((((.((((..((....))-------..)))).))))))))))....))).(((.((..(((.........)))..)).))--)..... ( -26.90) >DroYak_CAF1 10928 94 - 1 CCCACUUGGCACACAUUUCCAUUCCGCUUUUGC-------UAUAU--GGGUGUGUGCACCUGUGUACCUGCCUGUAUCUACAAUUUAUCUGCCGGUGUUUGCC ..(((...((((((((..((((...((....))-------...))--))))))))))....)))((((.((..(((.........)))..)).))))...... ( -26.20) >consensus CCCACUUGGCACACAUUCCC_____CCUUUUGC_______UAUAU__GGGUGUGUGCACCUGUGUACCUGUCUGUAUCUACAAUUUAUCUGCCGG__UUUGGC ..(((...((((((((((.............................))))))))))....)))..((.((..(((.........)))..)).))........ (-15.90 = -15.65 + -0.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:55 2006