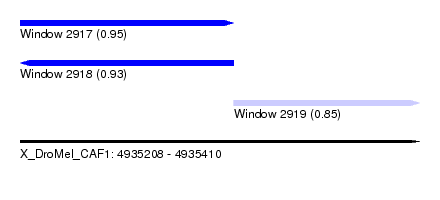
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,935,208 – 4,935,410 |
| Length | 202 |
| Max. P | 0.953970 |

| Location | 4,935,208 – 4,935,316 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.43 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -25.04 |
| Energy contribution | -24.43 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.953970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4935208 108 + 22224390 ------AAACCGUG---C---ACCAAAAUGGCGGACAUCAUGCGUCCGCCGUACAGUGAGAUCAAACGGCCGGACGAAAUCGUCAGAAUGACAACCGCCGACAAUCUCAAGUUCGCCGUC ------....((.(---(---......(((((((((.......)))))))))....((((((....((((.((..(....)(((.....)))..))))))...)))))).....)))).. ( -36.00) >DroVir_CAF1 9402 104 + 1 CUCG-------------A---UACAAAAUGGCGGACAUCAUGCGUCCGCCGUUUAGCGACAUUAAGCGGCCCGACGAAAUUGUCAGAAUGACGACCGCCGACAAUCUCAAAUUUGCCGUC .(((-------------.---....(((((((((((.......)))))))))))..)))......(((((..((((....))))(((.((.((.....)).)).))).......))))). ( -31.50) >DroGri_CAF1 11855 108 + 1 CUCCAAAA---------A---UACAAAAUGGCGGACAUCAUGCGUCCGCCGUACAGCGAUAUAAAGCGGCCCGACGAAAUUGUCAGAAUGACAACCGCCGAUAAUUUGAAAUUCGCCGUA ........---------.---......(((((((((.......)))))))))((.((((..((((.((((.........(((((.....)))))..))))....))))....)))).)). ( -30.20) >DroSim_CAF1 2 95 + 1 -------------------------AAAUGGCGGACAUCAUGCGUCCGCCGUACAGUGAGAUCAAACGGCCGGACAAAAUCGUCAAAAUGACAACCGCCGACAAUCUCAAGUUCGCCGUC -------------------------..(((((((((.....(((((((((((....((....)).)))).)))))......(((.....)))....)).((.....))..))))))))). ( -32.30) >DroWil_CAF1 8230 117 + 1 CAC---AAAAACUUCAACGCAAACAAAAUGGCGGACAUCAUGCGUCCGCCGUUUAGUGAGAUCAAACGGCCGGAUGAGAUUGUCAGAAUGACAACCGCCGAUAACUUGAAAUUUGCCGUC ...---............((((((.(((((((((((.......))))))))))).)).........((((.((......(((((.....)))))))))))............)))).... ( -33.70) >DroMoj_CAF1 12889 104 + 1 CUCG-------------U---UACAAAAUGGCGGACAUCAUGCGUCCGCCGUUCAGCGACAUUAAGCGGCCCGACGAAAUCGUCAGAAUGACAACCGCCGAUAAUCUGAAAUUUGCCGUC ...(-------------(---..(..((((((((((.......))))))))))..)..)).....(((((..((((....))))(((.((.(.......).)).))).......))))). ( -31.80) >consensus CUC______________A___UACAAAAUGGCGGACAUCAUGCGUCCGCCGUACAGCGAGAUCAAACGGCCCGACGAAAUCGUCAGAAUGACAACCGCCGACAAUCUCAAAUUCGCCGUC ...........................(((((((((.......))))))))).............(((((..((((....)))).((((.....................))))))))). (-25.04 = -24.43 + -0.61)

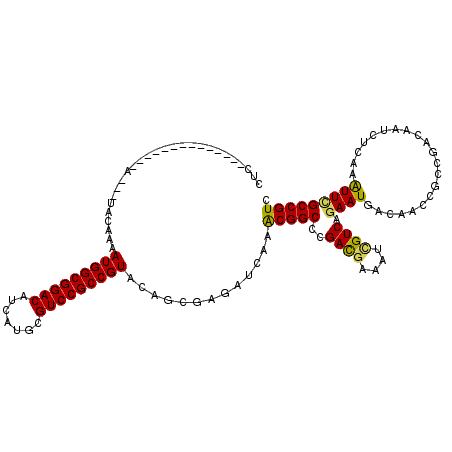

| Location | 4,935,208 – 4,935,316 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.43 |
| Mean single sequence MFE | -37.59 |
| Consensus MFE | -27.69 |
| Energy contribution | -27.25 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4935208 108 - 22224390 GACGGCGAACUUGAGAUUGUCGGCGGUUGUCAUUCUGACGAUUUCGUCCGGCCGUUUGAUCUCACUGUACGGCGGACGCAUGAUGUCCGCCAUUUUGGU---G---CACGGUUU------ ..(((((.((.((((((((((((((((((((.....))))))..)).))))).....)))))))..)).)((((((((.....))))))))........---)---).))....------ ( -40.30) >DroVir_CAF1 9402 104 - 1 GACGGCAAAUUUGAGAUUGUCGGCGGUCGUCAUUCUGACAAUUUCGUCGGGCCGCUUAAUGUCGCUAAACGGCGGACGCAUGAUGUCCGCCAUUUUGUA---U-------------CGAG ((..(((((...(((((((((((..(....)...)))))))))))((..(((.((.....)).)))..))((((((((.....))))))))..))))).---)-------------)... ( -36.70) >DroGri_CAF1 11855 108 - 1 UACGGCGAAUUUCAAAUUAUCGGCGGUUGUCAUUCUGACAAUUUCGUCGGGCCGCUUUAUAUCGCUGUACGGCGGACGCAUGAUGUCCGCCAUUUUGUA---U---------UUUUGGAG ((((((((...........((((((((((((.....))))))..))))))...........)))))))).((((((((.....))))))))........---.---------........ ( -38.65) >DroSim_CAF1 2 95 - 1 GACGGCGAACUUGAGAUUGUCGGCGGUUGUCAUUUUGACGAUUUUGUCCGGCCGUUUGAUCUCACUGUACGGCGGACGCAUGAUGUCCGCCAUUU------------------------- .((((.((...(.((((.(((((((((((((.....))))))..)).))))).)))).)..)).))))..((((((((.....))))))))....------------------------- ( -35.40) >DroWil_CAF1 8230 117 - 1 GACGGCAAAUUUCAAGUUAUCGGCGGUUGUCAUUCUGACAAUCUCAUCCGGCCGUUUGAUCUCACUAAACGGCGGACGCAUGAUGUCCGCCAUUUUGUUUGCGUUGAAGUUUUU---GUG .((((.(((((((((......((((((((((.....))))))).......((((((((.......))))))))(((((.....)))))))).....(....).)))))))))))---)). ( -36.60) >DroMoj_CAF1 12889 104 - 1 GACGGCAAAUUUCAGAUUAUCGGCGGUUGUCAUUCUGACGAUUUCGUCGGGCCGCUUAAUGUCGCUGAACGGCGGACGCAUGAUGUCCGCCAUUUUGUA---A-------------CGAG ..(((((((.(((((..(((.((((((.......((((((....))))))))))))..)))...))))).((((((((.....))))))))..))))).---.-------------)).. ( -37.91) >consensus GACGGCAAAUUUCAGAUUAUCGGCGGUUGUCAUUCUGACAAUUUCGUCCGGCCGCUUGAUCUCACUGAACGGCGGACGCAUGAUGUCCGCCAUUUUGUA___G______________GAG ..((((..((........)).((((((((((.....))))))..))))..))))................((((((((.....))))))))............................. (-27.69 = -27.25 + -0.44)

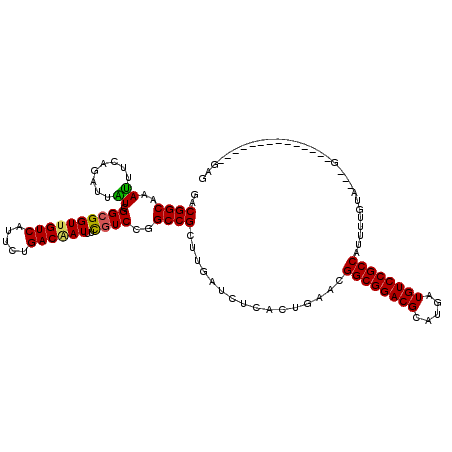
| Location | 4,935,316 – 4,935,410 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 75.41 |
| Mean single sequence MFE | -29.77 |
| Consensus MFE | -19.59 |
| Energy contribution | -19.57 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4935316 94 + 22224390 CUCAUUGGCCUCAUCGAGGUUGGCCAGGUGACCAACCGGGAGGUGGUCAACACCGUUCUGCAUUUGGUAAGUGCUAGU---UACAGCCAAUUAGCCG ......((((((...)))((((((..((((((((.((....)))))))...)))((.(((((((.....))))).)).---.)).))))))..))). ( -31.00) >DroGri_CAF1 11963 74 + 1 CUCAUCGGCCUCAUCGAGGUUGGCCAGGUGACCAAUCGGGAGGUGGUCAACACCGUGUUACAUUUGGUAAGUGU----------------------- .....(((((((...)))))))((((((((.((....))..((((.....))))......))))))))......----------------------- ( -25.30) >DroEre_CAF1 9622 93 + 1 CUCAUCGGCCUCAUCGAGGUUGGCCAGGUGACCAACCGGGAGGUGGUGAACACCGUUCUGCAUUUGGUAAGUGCCAGC---AGC-GCUAGUCAGCCG ....((((.....))))(((((((..((((((((.((....))))))...))))((.((((...((((....))))))---)).-))..))))))). ( -35.00) >DroWil_CAF1 8347 92 + 1 CUCAUCGGCCUAAUCGAGGUGGGCCAGGUGACCAAUCGUGAGGUGGUCAAUACUGUGUUACAUUUGGUAAGUACCCAUAUGAAUAA-----CUACUU ......((((((.......))))))((.((((((.((....))))))))...))(((((((((.(((.......))).)))..)))-----).)).. ( -26.70) >DroYak_CAF1 9092 77 + 1 CUCAUCGGCCUUAUCGAGGUUGGCCAGGUGACCAACCGGGAGGUGGUCAACACCGUGCUGCAUUUGGUAAGUGCCAC-------------------- ......((((((((((((...(((..((((((((.((....)))))))...)))..)))...))))))))).)))..-------------------- ( -32.20) >DroPer_CAF1 8575 91 + 1 CUCAUCGGCCUCAUCGAGGUGGGGCAGGUCACCAACCGCGAGGUUGUGAACACCGUACUGCAUUUGGUAAGUGCUAUU---UAUAA---AAUAAUCU .......(((((((....))))))).((((((.((((....))))))))...))(((((((.....)).)))))....---.....---........ ( -28.40) >consensus CUCAUCGGCCUCAUCGAGGUUGGCCAGGUGACCAACCGGGAGGUGGUCAACACCGUGCUGCAUUUGGUAAGUGCCAGU____A_________A_CC_ ......(((((.((((((((.(..(.(.((((((.((....))))))))...).)..).)).)))))).)).)))...................... (-19.59 = -19.57 + -0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:52 2006