
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,923,765 – 4,923,939 |
| Length | 174 |
| Max. P | 0.975630 |

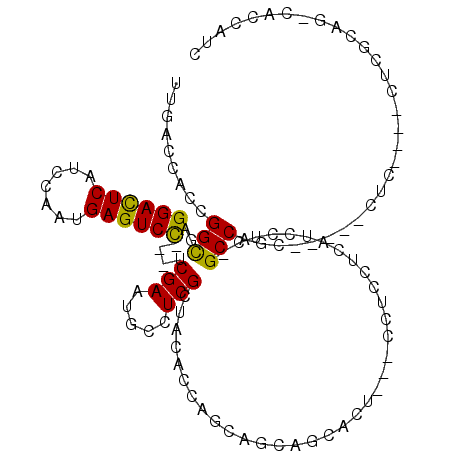
| Location | 4,923,765 – 4,923,862 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.67 |
| Mean single sequence MFE | -25.94 |
| Consensus MFE | -13.08 |
| Energy contribution | -12.50 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4923765 97 - 22224390 UUGACCACCGGCGAGGACUCAUCG---GAGUCC---UCGAAUGCCUCGCUACACCUGAACCAGCACUCCUCCACCUCAUCCUC-GCCAGC------------CUCGCAG-CACCAAG (((......((((((((......(---(((...---..((.....))(((...........)))....))))......)))))-))).((------------......)-)..))). ( -23.50) >DroPse_CAF1 11976 107 - 1 UUGACCACCGGUGAGGAUUCAUCCAAUGAGUCC---UCGAAUGCCUCGCUACACAAUCAGCCGCACU---CCUCCUCAUCCGCAGCCAGCAUCAGCUC----CUCGCAUUCACCAUC .........(((((((((((((...))))))))---))((((((...............((.((...---...........)).)).(((....))).----...)))))))))... ( -28.44) >DroEre_CAF1 10209 97 - 1 UUGACCACCGGCGAGGACUCAUCG---GAGUCC---UCGAAUGCCUCGCUACACCUGAACCAGCACUCCUCCACCUCAUCCUC-GCCAGC------------CUCGCAG-CACCAUG .........((((((((......(---(((...---..((.....))(((...........)))....))))......)))))-))).((------------......)-)...... ( -23.40) >DroMoj_CAF1 15265 92 - 1 UUAACCACCGGCGAGGACUCCUCCAAUGAGUCCUCAUCGAAUGCCUCGCUACAGCA---GCCGCACU------CCUCA------GCCAG------CUC----AACGCACUCACAGCA .........(((((((((((.......))))))))...((.(((...(((.....)---)).))).)------)....------))).(------(..----...)).......... ( -25.70) >DroAna_CAF1 10913 110 - 1 UUGACCACCGGCGAGGACUCAUCCAAUGAGUCU---UCGAAUGCCUCGCUCCACCAGCAGCAUCACUCCUCGUCCUCAUCCUC-GCCAGCAACGCAUUCAAACUCGCAA-CACCA-- ..........((((((((((((...))))))).---..((((((...(((.....))).((......................-....))...))))))...)))))..-.....-- ( -26.17) >DroPer_CAF1 11288 107 - 1 UUGACCACCGGUGAGGAUUCAUCCAAUGAGUCC---UCGAAUGCCUCGCUACACAAUCAGCCGCACU---CCUCCUCAUCCGCAGCCAGCAUCAGCUC----CUCGCAUUCACCAUC .........(((((((((((((...))))))))---))((((((...............((.((...---...........)).)).(((....))).----...)))))))))... ( -28.44) >consensus UUGACCACCGGCGAGGACUCAUCCAAUGAGUCC___UCGAAUGCCUCGCUACACCAGCAGCAGCACU___CCUCCUCAUCCUC_GCCAGC_____CUC____CUCGCAG_CACCAUC .........(((..((((((.......))))))....(((.....)))....................................))).............................. (-13.08 = -12.50 + -0.58)


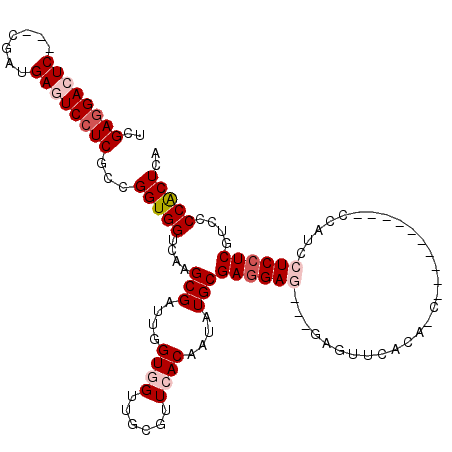
| Location | 4,923,828 – 4,923,939 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.97 |
| Mean single sequence MFE | -38.83 |
| Consensus MFE | -27.12 |
| Energy contribution | -28.20 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4923828 111 + 22224390 UCGAGGACUC---CGAUGAGUCCUCGCCGGUGGUCAAGCGAUUGGUGGUUGCGUUCACAAUAUGCGAGGAG---GAGUUCAUACCGCCGCCGCUGCCAUCCUCCUCGUCCCCGCUCA .(((((((((---....))))))))).(((.((.(..(((....((((......))))....)))((((((---((...((....((....))))...))))))))).))))).... ( -43.70) >DroPse_CAF1 12046 96 + 1 UCGAGGACUCAUUGGAUGAAUCCUCACCGGUGGUCAAGCGAUUGGUGGUUGAGUUGACAAUAUGCGAGGAA---GAGUUCACA------------------UCCUCGUCCCCACUGA ..(((((.((((...)))).)))))..((((((((((.((((.....))))..))))).....(((((((.---(......).------------------)))))))...))))). ( -32.40) >DroSec_CAF1 10476 111 + 1 UCGAGGACUC---CGAUGAGUCCUCGCCGGUGGUCAAGCGAUUGGUGGUUGCGUUCACAAUAUGCGAGGAG---GAGUUCAUACCGCCGCCGCUGCCAUCCUCCUCGUCCCCGCUCA .(((((((((---....))))))))).(((.((.(..(((....((((......))))....)))((((((---((...((....((....))))...))))))))).))))).... ( -43.70) >DroYak_CAF1 10237 111 + 1 UCGAGGACUC---CGAUGAGUCCUCGCCGGUGGUCAAGCGAUUGGUGGUUGCGUUCACAAUAUGCGAGGAG---GAAUUCAUACCGCCGCCGCUGCCAUCCUCCUCGUCCCCGCUCA .(((((((((---....))))))))).(((.((.(..(((....((((......))))....)))((((((---((...((....((....))))...))))))))).))))).... ( -42.60) >DroAna_CAF1 10986 105 + 1 UCGAAGACUCAUUGGAUGAGUCCUCGCCGGUGGUCAAGCGAUUGGUGGUUGCGUUCACGAUGUGCGAUGAGGAUGAGUUCAC--C----------AUAUCCUCCUCAUCCCCACUGC .(((.(((((((...))))))).))).((((((....(((..(.((((......)))).)..)))((((((((.((......--.----------...)).)))))))).)))))). ( -38.20) >DroPer_CAF1 11358 96 + 1 UCGAGGACUCAUUGGAUGAAUCCUCACCGGUGGUCAAGCGAUUGGUGGUUGAGUUGACAAUAUGCGAGGAA---GAGUUCACA------------------UCCUCGUCCCCACUGA ..(((((.((((...)))).)))))..((((((((((.((((.....))))..))))).....(((((((.---(......).------------------)))))))...))))). ( -32.40) >consensus UCGAGGACUC___CGAUGAGUCCUCGCCGGUGGUCAAGCGAUUGGUGGUUGCGUUCACAAUAUGCGAGGAG___GAGUUCACA_C__________CCAUCCUCCUCGUCCCCACUCA ..((((((((.......))))))))...(((((....(((....((((......))))....)))((((((.............................))))))....))))).. (-27.12 = -28.20 + 1.08)


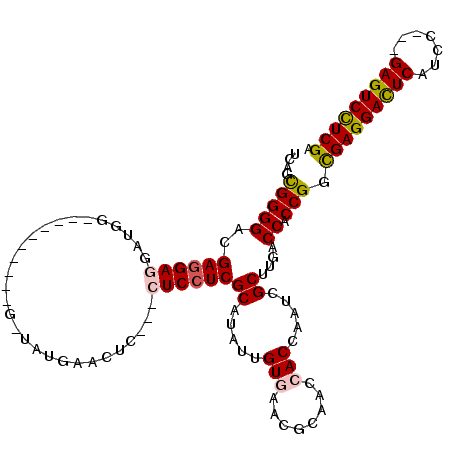
| Location | 4,923,828 – 4,923,939 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.97 |
| Mean single sequence MFE | -38.98 |
| Consensus MFE | -27.47 |
| Energy contribution | -27.47 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4923828 111 - 22224390 UGAGCGGGGACGAGGAGGAUGGCAGCGGCGGCGGUAUGAACUC---CUCCUCGCAUAUUGUGAACGCAACCACCAAUCGCUUGACCACCGGCGAGGACUCAUCG---GAGUCCUCGA ....(((((.((((((((.(((..(((..((.((........)---).))((((.....)))).)))..)))))..)).)))).)).))).(((((((((....---))))))))). ( -44.90) >DroPse_CAF1 12046 96 - 1 UCAGUGGGGACGAGGA------------------UGUGAACUC---UUCCUCGCAUAUUGUCAACUCAACCACCAAUCGCUUGACCACCGGUGAGGAUUCAUCCAAUGAGUCCUCGA ((.((((.(((((..(------------------(((((....---....)))))).)))))...((((.(.......).)))))))).))(((((((((((...))))))))))). ( -31.00) >DroSec_CAF1 10476 111 - 1 UGAGCGGGGACGAGGAGGAUGGCAGCGGCGGCGGUAUGAACUC---CUCCUCGCAUAUUGUGAACGCAACCACCAAUCGCUUGACCACCGGCGAGGACUCAUCG---GAGUCCUCGA ....(((((.((((((((.(((..(((..((.((........)---).))((((.....)))).)))..)))))..)).)))).)).))).(((((((((....---))))))))). ( -44.90) >DroYak_CAF1 10237 111 - 1 UGAGCGGGGACGAGGAGGAUGGCAGCGGCGGCGGUAUGAAUUC---CUCCUCGCAUAUUGUGAACGCAACCACCAAUCGCUUGACCACCGGCGAGGACUCAUCG---GAGUCCUCGA .(((((.((.(((((((((...((...((....)).))...))---)))))))....((((....))))...))...))))).........(((((((((....---))))))))). ( -45.20) >DroAna_CAF1 10986 105 - 1 GCAGUGGGGAUGAGGAGGAUAU----------G--GUGAACUCAUCCUCAUCGCACAUCGUGAACGCAACCACCAAUCGCUUGACCACCGGCGAGGACUCAUCCAAUGAGUCUUCGA ((.((((.((((((((......----------(--(....))..))))))))((.....(((........))).....))....))))..))((((((((((...)))))))))).. ( -36.90) >DroPer_CAF1 11358 96 - 1 UCAGUGGGGACGAGGA------------------UGUGAACUC---UUCCUCGCAUAUUGUCAACUCAACCACCAAUCGCUUGACCACCGGUGAGGAUUCAUCCAAUGAGUCCUCGA ((.((((.(((((..(------------------(((((....---....)))))).)))))...((((.(.......).)))))))).))(((((((((((...))))))))))). ( -31.00) >consensus UCAGCGGGGACGAGGAGGAUGG__________G_UAUGAACUC___CUCCUCGCAUAUUGUGAACGCAACCACCAAUCGCUUGACCACCGGCGAGGACUCAUCC___GAGUCCUCGA ....(((((..((((((.............................))))))((.....(((........))).....))....)).))).(((((((((.......))))))))). (-27.47 = -27.47 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:50 2006