
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,890,313 – 4,890,445 |
| Length | 132 |
| Max. P | 0.772969 |

| Location | 4,890,313 – 4,890,412 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 86.51 |
| Mean single sequence MFE | -28.21 |
| Consensus MFE | -21.02 |
| Energy contribution | -21.47 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
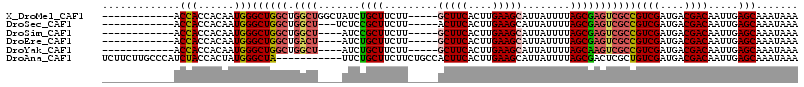
>X_DroMel_CAF1 4890313 99 + 22224390 ------------ACCACCACAAUGGGCUGGCUGGCUGGCUAUCUGCUUCUU-----GCUUCACUUGAAGCAUUAUUUUAGCGAGUCGCCGUCGAUGACGACAAUUGAGCAAAUAAA ------------.(((......)))((((((.((((.((((.........(-----(((((....))))))......)))).)))))))((((....)))).....)))....... ( -33.16) >DroSec_CAF1 41489 96 + 1 ------------ACCACCACAAUGGGCUGGCUGGCU---UCUCCGCUUCUU-----ACUUCACUUGAAGCAUUAUUUUAGCGAGUCGCCGUCGAUGACGACAAUUGAGCAAAUAAA ------------.......((((.(((.((((.(((---.....(((((..-----.........)))))........))).)))))))((((....)))).)))).......... ( -25.62) >DroSim_CAF1 42708 95 + 1 ------------ACCACCACAAUGGGCUGGCUGGCU----AUCCGCUUCUU-----GCUUCACUUGAAGCAUUAUUUUAGCGAGUCGCCGUCGAUGACGACAAUUGAGCAAAUAAA ------------.......((((.(((.((((.(((----(.........(-----(((((....))))))......)))).)))))))((((....)))).)))).......... ( -29.66) >DroEre_CAF1 44315 95 + 1 ------------ACCACCACAAUGGGCUGGCUGACU----AUCUGCUUCUU-----GCUUCACUUGAAGCAUUAUUUUAGCGAGUCGCCGUCGAUGACGACAAUUGAGCAAAUAAA ------------.(((......)))((((((.((((----...((((...(-----(((((....)))))).......)))))))))))((((....)))).....)))....... ( -28.60) >DroYak_CAF1 42296 95 + 1 ------------ACCACCACAAUGGGCUGGCUGGCU----AUCUGCUUCUU-----GCUUCACUUGAAGCAUUAUUUUAGCAAGUCGCCGUCGAUGACGACAAUUGAGCAAAUAAA ------------.(((......)))((((((.((((----...((((...(-----(((((....)))))).......)))))))))))((((....)))).....)))....... ( -28.70) >DroAna_CAF1 101116 105 + 1 UCUUCUUGCCCAUCUACCACUAUGGGCUA-----------UUCUGCUUCUUCUGCCACUUCACUUGAAGCAUUAUUUUAGCGACUCGCUGUCGAUGACGACAAUUGAGCAAAUAAA .......((((((........))))))..-----------...((((((...((......))...))))))..........(.((((.(((((....)))))..)))))....... ( -23.50) >consensus ____________ACCACCACAAUGGGCUGGCUGGCU____AUCUGCUUCUU_____GCUUCACUUGAAGCAUUAUUUUAGCGAGUCGCCGUCGAUGACGACAAUUGAGCAAAUAAA .............(((......)))((((((.((((.......((((.........(((((....)))))........)))))))))))((((....)))).....)))....... (-21.02 = -21.47 + 0.45)
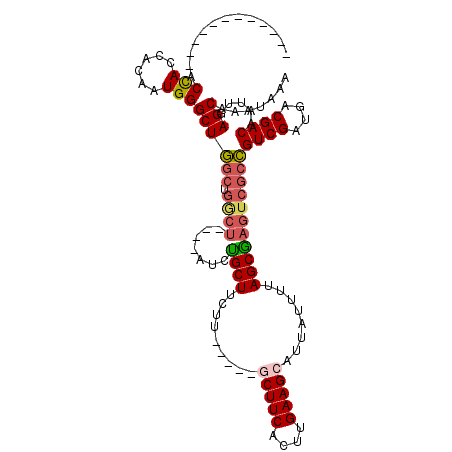
| Location | 4,890,340 – 4,890,445 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 83.11 |
| Mean single sequence MFE | -25.28 |
| Consensus MFE | -18.29 |
| Energy contribution | -18.02 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4890340 105 - 22224390 UUUUUUUAUUACAACUUUU------UGUUG-GUUCUUAUCUUUAUUUGCUCAAUUGUCGUCAUCGACGGCGACUCGCUAAAAUAAUGCUUCAAGUGAAGC-----AAGAAGCAGAUA ...........((((....------.))))-...........(((((((((..((((((((...)))))))).............((((((....)))))-----).).)))))))) ( -26.90) >DroSec_CAF1 41513 97 - 1 --------UUACAACUCUU------UUUUG-GUUCUUAUCUUUAUUUGCUCAAUUGUCGUCAUCGACGGCGACUCGCUAAAAUAAUGCUUCAAGUGAAGU-----AAGAAGCGGAGA --------..........(------(((((-.((((((.(((((((((.....((((((((...))))))))...((.........))..))))))))))-----))))).)))))) ( -26.80) >DroSim_CAF1 42732 97 - 1 --------UUACAACUCUU------UUUUGGGUUCUUAUCUUUAUUUGCUCAAUUGUCGUCAUCGACGGCGACUCGCUAAAAUAAUGCUUCAAGUGAAGC-----AAGAAGCGGAU- --------....(((((..------....))))).........((((((((..((((((((...)))))))).............((((((....)))))-----).).)))))))- ( -24.60) >DroEre_CAF1 44339 96 - 1 --------CUACUACCUUU------UUUUG-GUUCUUAUCUUUAUUUGCUCAAUUGUCGUCAUCGACGGCGACUCGCUAAAAUAAUGCUUCAAGUGAAGC-----AAGAAGCAGAU- --------...........------.((((-.(((((....((((((((....((((((((...))))))))...))..)))))).(((((....)))))-----))))).)))).- ( -24.30) >DroYak_CAF1 42320 100 - 1 --------UU--AACCUUUUUUUUAUUUUG-GUUCUUAUCUUUAUUUGCUCAAUUGUCGUCAUCGACGGCGACUUGCUAAAAUAAUGCUUCAAGUGAAGC-----AAGAAGCAGAU- --------..--..............((((-.(((((....((((((...(((((((((((...)))))))).)))...)))))).(((((....)))))-----))))).)))).- ( -26.30) >DroAna_CAF1 101145 100 - 1 -UGUUUUAG--------UG------UUGUG-UUUUUUAUCUUUAUUUGCUCAAUUGUCGUCAUCGACAGCGAGUCGCUAAAAUAAUGCUUCAAGUGAAGUGGCAGAAGAAGCAGAA- -((((((((--------((------.....-................((((..((((((....)))))).)))))))))))))).((((((...((......))...))))))...- ( -22.79) >consensus ________UUACAACUUUU______UUUUG_GUUCUUAUCUUUAUUUGCUCAAUUGUCGUCAUCGACGGCGACUCGCUAAAAUAAUGCUUCAAGUGAAGC_____AAGAAGCAGAU_ ............................................(((((((..((((((((...))))))))..............(((((....))))).......).)))))).. (-18.29 = -18.02 + -0.28)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:36 2006