
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,885,309 – 4,885,432 |
| Length | 123 |
| Max. P | 0.873089 |

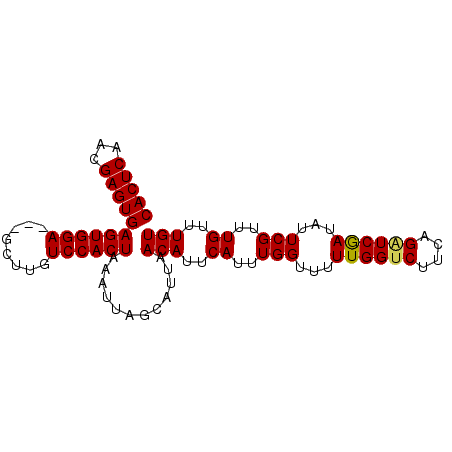
| Location | 4,885,309 – 4,885,400 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 96.28 |
| Mean single sequence MFE | -21.32 |
| Consensus MFE | -20.06 |
| Energy contribution | -20.00 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4885309 91 + 22224390 CACUCAACGAGUGAGUGGAGUAGCUUGUCCACUAAAUUAGCAUUAACAUUCAUUUGGUUUUUGGUCUUCAGAUCGAUAUUCGUUUGUUUGU (((((...)))))((((((........))))))............(((..((..(((...((((((....))))))...)))..))..))) ( -22.10) >DroSim_CAF1 37559 88 + 1 CACUCAACGAGUGAGUGGA---GCUUGUCCACUAAAUUAGCAUUAACAUUCAUUUGGUUUUUGGUCUUCAGAUCAAAAUUCGUUUGUUUGU (((((...)))))((((((---.....))))))............(((..((..((..((((((((....))))))))..))..))..))) ( -24.60) >DroEre_CAF1 39486 88 + 1 CACUCAACGAGUGAGUGGA---GCUUGUCCACUAAAUUAGCAUUAACAUUCAUUUGGUUUUUGGUCUUCAGGUCGAUAUUCGUUUGUUUGU (((((...)))))((((((---.....))))))............(((..((..(((...(((..(....)..)))...)))..))..))) ( -20.90) >DroYak_CAF1 37363 88 + 1 CACUCAACGAGUGAGUGGA---GCUUGUCCACUAAAUUAGCAUUAACAUUCAUUUGGUUUUUGGUCUUCAGUUCGAUAUUCGUUUGUUUGU (((((...)))))((((((---.....))))))............(((..((..(((...((((........))))...)))..))..))) ( -17.70) >consensus CACUCAACGAGUGAGUGGA___GCUUGUCCACUAAAUUAGCAUUAACAUUCAUUUGGUUUUUGGUCUUCAGAUCGAUAUUCGUUUGUUUGU (((((...)))))((((((........))))))............(((..((..(((...((((((....))))))...)))..))..))) (-20.06 = -20.00 + -0.06)


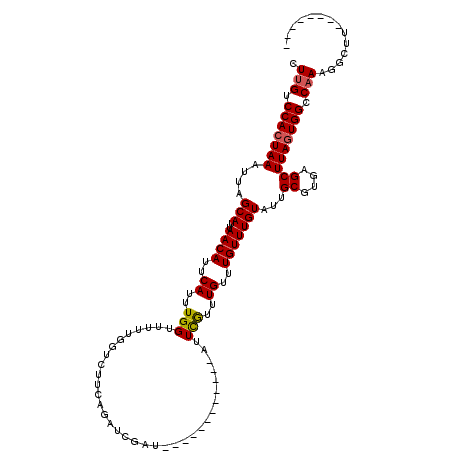
| Location | 4,885,332 – 4,885,432 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.80 |
| Mean single sequence MFE | -27.06 |
| Consensus MFE | -16.26 |
| Energy contribution | -16.34 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4885332 100 + 22224390 CUUGUCCACUAAAUUAGCAUUAACAUUCAUUUGGUUUUUGGUCUUCAGAUCGAU------------AUUCGUUUGUUUGUUUGUAUUGCGUGAGCUUAGUGGCCAAAGGCUU-------- .(((.(((((((....(((..((((..((..(((...((((((....)))))).------------..)))..))..)))))))...((....))))))))).)))......-------- ( -27.90) >DroSim_CAF1 37579 100 + 1 CUUGUCCACUAAAUUAGCAUUAACAUUCAUUUGGUUUUUGGUCUUCAGAUCAAA------------AUUCGUUUGUUUGUUUGUGUUGCGUGAGCUUAGUGGCCAAAGGCUU-------- .(((.(((((((...(((((.((((..((..((..((((((((....)))))))------------)..))..))..)))).)))))((....))))))))).)))......-------- ( -34.40) >DroEre_CAF1 39506 100 + 1 CUUGUCCACUAAAUUAGCAUUAACAUUCAUUUGGUUUUUGGUCUUCAGGUCGAU------------AUUCGUUUGUUUGUUUGUAUUGCGUGAGCUUAGUGGCCAAAGGCUU-------- .(((.(((((((....(((..((((..((..(((...(((..(....)..))).------------..)))..))..)))))))...((....))))))))).)))......-------- ( -26.70) >DroYak_CAF1 37383 108 + 1 CUUGUCCACUAAAUUAGCAUUAACAUUCAUUUGGUUUUUGGUCUUCAGUUCGAU------------AUUCGUUUGUUUGUUUGUAUUGCGUGAGCUUAGUGGCCAAAGGCUUAGUGGCCA .(((.(((((((....(((..((((..((..(((...((((........)))).------------..)))..))..)))))))...((....))))))))).))).((((....)))). ( -29.40) >DroAna_CAF1 94984 101 + 1 CUUGUCCAGUAAAUUAGCAUUAACAUUCAUUUGGUUU--------CAGACCGAUUCGUUUCGUUUCGUUUAUUUGUUUGUUUGUAUUGCGUGAGCUUAGUGGCCCAAAC----------- .(((.(((.(((....(((..((((..((...((((.--------..))))((..((...))..)).......))..)))))))...((....))))).)))..)))..----------- ( -16.90) >consensus CUUGUCCACUAAAUUAGCAUUAACAUUCAUUUGGUUUUUGGUCUUCAGAUCGAU____________AUUCGUUUGUUUGUUUGUAUUGCGUGAGCUUAGUGGCCAAAGGCUU________ .(((.(((((((.((((((..((((..((..(((..................................)))..))..)))).....))).)))..))))))).))).............. (-16.26 = -16.34 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:31 2006