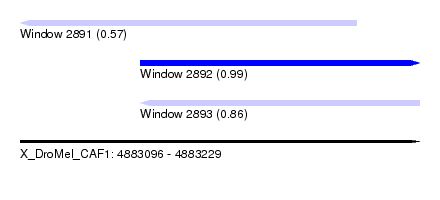
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,883,096 – 4,883,229 |
| Length | 133 |
| Max. P | 0.989919 |

| Location | 4,883,096 – 4,883,208 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.28 |
| Mean single sequence MFE | -26.55 |
| Consensus MFE | -17.16 |
| Energy contribution | -18.72 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
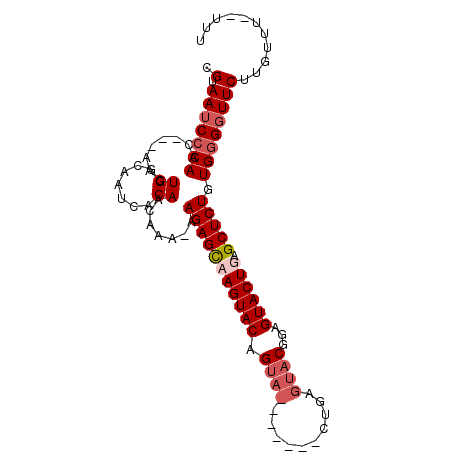
>X_DroMel_CAF1 4883096 112 - 22224390 UAAUCACAACAAAAAAGAGUCAGUACAGUA-------CUGAGUACUGAGUACUGAGCUCUGUGGGGUUCUUGUUU-UUUUGUUAUCUUAUUCGACUAAUACUUGUUCUUUUUCUACAAAA .......(((((((((((.(((((((....-------....))))))).)...((((((....))))))...)))-)))))))..................................... ( -22.40) >DroSim_CAF1 35493 120 - 1 CAAUCACAACAAAAAAGAGCCAGUACAGUACAGUACACUGAGUACUGAGUACUGAGCUCUGUGGGUUUCUUGUUUUUUUUGUUAUCUUAUUCGACUAAUACUUGUUCUUUUUCUACAAAA .......(((((((((((((((((((....((((((.....)))))).)))))).)))))(..((...))..)..))))))))..................................... ( -26.30) >DroEre_CAF1 37213 103 - 1 CAAUCACAACAAA-AAGAGCAAGUACAG--------------UACGGAGUACUGAGCUCUGUGGGGUUCUUGUUU--UUUGUUAUCUUAUUCGACUAAUACUUGUUCUUUUUCUACAAAA ..........(((-(((((((((((.((--------------(((((((.((.((((((....))))))..)).)--)))))...........)))..))))))))))))))........ ( -24.80) >DroYak_CAF1 35077 117 - 1 CAAUCACAACAAA-AAGAGCAAGUACAGUACUGAGUACUCAGUACGGAGUACUGAGCUCUGUGGGGUUCUUGUUU--UUUGUUAUCUUAUUCGACUAAUACUUGUUCUUUUUCUACAAAA ..........(((-(((((((((((..(((((((....)))))))((((.((.((((((....))))))..)).)--)))..................))))))))))))))........ ( -32.70) >consensus CAAUCACAACAAA_AAGAGCAAGUACAGUA_______CUGAGUACGGAGUACUGAGCUCUGUGGGGUUCUUGUUU__UUUGUUAUCUUAUUCGACUAAUACUUGUUCUUUUUCUACAAAA ...........((((((((((((((...............(((((...)))))((((((....)))))).............................))))))))))))))........ (-17.16 = -18.72 + 1.56)

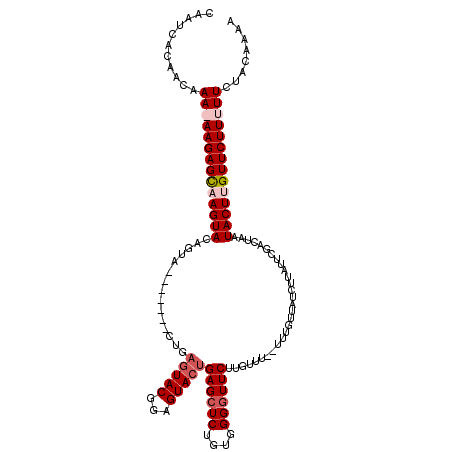
| Location | 4,883,136 – 4,883,229 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 82.74 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -18.33 |
| Energy contribution | -20.20 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4883136 93 + 22224390 AAAA-AAACAAGAACCCCACAGAGCUCAGUACUCAGUACUCAG-------UACUGUACUGACUCUUUUUUGUUGUGAUUACCAGUUGUUGUUGGGAUUACG ....-.((((((((......((((.(((((((..(((((...)-------)))))))))))))))))))))))((((((.((((......)))))))))). ( -27.40) >DroSim_CAF1 35533 101 + 1 AAAAAAAACAAGAAACCCACAGAGCUCAGUACUCAGUACUCAGUGUACUGUACUGUACUGGCUCUUUUUUGUUGUGAUUGCCAGUAGUUGUUGCGAUUACG ......((((((((......(((((.((((((.((((((.....))))))....)))))))))))))))))))((((((((...........)))))))). ( -35.40) >DroEre_CAF1 37253 81 + 1 AAA--AAACAAGAACCCCACAGAGCUCAGUACUCCGUA--------------CUGUACUUGCUCUU-UUUGUUGUGAUUGCCAGUU---GUUGGGAUUACG ...--.(((((((.......(((((..(((((......--------------..))))).))))))-))))))((((((.((((..---.)))))))))). ( -20.71) >DroYak_CAF1 35117 95 + 1 AAA--AAACAAGAACCCCACAGAGCUCAGUACUCCGUACUGAGUACUCAGUACUGUACUUGCUCUU-UUUGUUGUGAUUGCCAGUA---GUUGGGAUUACG ...--.(((((((.......(((((..(((((...(((((((....))))))).))))).))))))-))))))((((((.((((..---.)))))))))). ( -30.21) >consensus AAA__AAACAAGAACCCCACAGAGCUCAGUACUCAGUACUCAG_______UACUGUACUGGCUCUU_UUUGUUGUGAUUGCCAGUA___GUUGGGAUUACG ......((((((((......((((.(((((((...((((..........)))).)))))))))))))))))))((((((.((((......)))))))))). (-18.33 = -20.20 + 1.87)

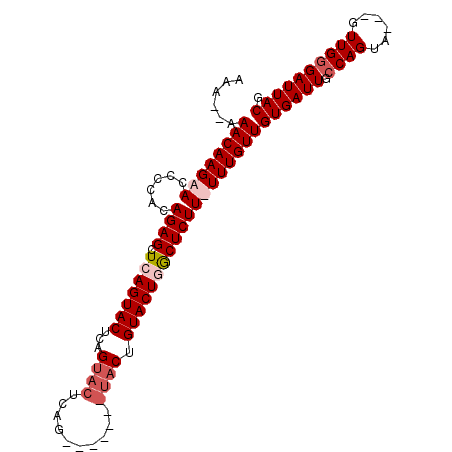

| Location | 4,883,136 – 4,883,229 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 82.74 |
| Mean single sequence MFE | -25.71 |
| Consensus MFE | -15.90 |
| Energy contribution | -17.52 |
| Covariance contribution | 1.63 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4883136 93 - 22224390 CGUAAUCCCAACAACAACUGGUAAUCACAACAAAAAAGAGUCAGUACAGUA-------CUGAGUACUGAGUACUGAGCUCUGUGGGGUUCUUGUUU-UUUU .(.(((((((........((.......)).......(((((((((((((((-------(...)))))..))))))).)))).))))))))......-.... ( -24.10) >DroSim_CAF1 35533 101 - 1 CGUAAUCGCAACAACUACUGGCAAUCACAACAAAAAAGAGCCAGUACAGUACAGUACACUGAGUACUGAGUACUGAGCUCUGUGGGUUUCUUGUUUUUUUU .........(((((..(((.(((..............((((((((((....((((((.....)))))).)))))).))))))).)))...)))))...... ( -26.33) >DroEre_CAF1 37253 81 - 1 CGUAAUCCCAAC---AACUGGCAAUCACAACAAA-AAGAGCAAGUACAG--------------UACGGAGUACUGAGCUCUGUGGGGUUCUUGUUU--UUU .......(((..---...)))............(-(((((((((.((..--------------((((((((.....))))))))..)).)))))))--))) ( -22.10) >DroYak_CAF1 35117 95 - 1 CGUAAUCCCAAC---UACUGGCAAUCACAACAAA-AAGAGCAAGUACAGUACUGAGUACUCAGUACGGAGUACUGAGCUCUGUGGGGUUCUUGUUU--UUU .(.(((((((..---...((.......)).....-.(((((.(((((.(((((((....)))))))...)))))..))))).))))))))......--... ( -30.30) >consensus CGUAAUCCCAAC___AACUGGCAAUCACAACAAA_AAGAGCAAGUACAGUA_______CUGAGUACGGAGUACUGAGCUCUGUGGGGUUCUUGUUU__UUU .(.(((((((........((.......)).......(((((((((((.((((..........))))...)))))).))))).))))))))........... (-15.90 = -17.52 + 1.63)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:29 2006