
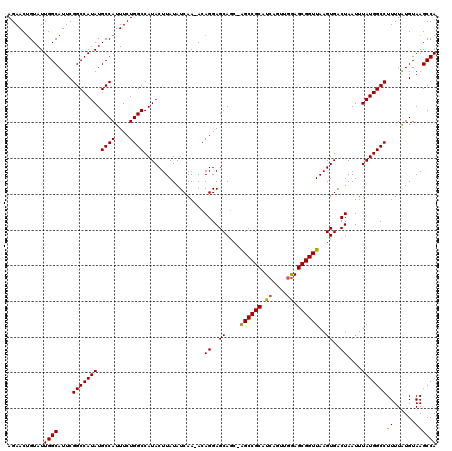
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,881,649 – 4,881,767 |
| Length | 118 |
| Max. P | 0.993656 |

| Location | 4,881,649 – 4,881,767 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.75 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -29.88 |
| Energy contribution | -29.77 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -0.84 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
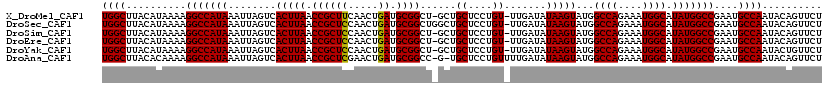
>X_DroMel_CAF1 4881649 118 + 22224390 AGAACUGUAUUGGCAUUCGGCCAUAUGCCAUUUCUGGCCAUACUUAUAUCAA-ACAGGAGCAGC-AGCCGCAUCAGUUGAAGCGGUUAAGUGACUAAUUUAUGGCCUUUUAUGUAAGCCA ..........((((....(((((((.((((....))))..............-..((..((...-((((((.((....)).))))))..))..))....)))))))((......)))))) ( -31.20) >DroSec_CAF1 32940 119 + 1 AGAACUGUAUUGGCAUUCGGCCAUAUGCCAUUUCUGGCCAUACUUAUAUCAA-ACAGGAGCAGCCAGCCGCAUCAGUUGGAGCGGUUAAGUGACUAAUUUAUGGCCUUUUAUGUAAGCCA ..........((((....(((.....)))......(((((((......(((.-...((.....))((((((..(....)..))))))...)))......)))))))..........)))) ( -32.00) >DroSim_CAF1 34123 118 + 1 AGAACUGUAUUGGCAUUCGGCCAUAUGCCAUUUCUGGCCAUACUUAUAUCAA-ACAGGAGCAGC-AGCCGCAUCAGUUGGAGCGGUUAAGUGACUAAUUUAUGGCCUUUUAUGUAAGCCA ..........((((....(((((((.((((....))))..............-..((..((...-((((((..(....)..))))))..))..))....)))))))((......)))))) ( -30.40) >DroEre_CAF1 35814 118 + 1 AGAACUGUAUUGGCAUUCGGCCAUAUGCCAUUUCUGGCCAUACUUAUAUCAA-ACAGGAGCAGC-AGCCGCAUCAGUUGGAGCGGUUAAGUGACUAAUUUAUGGCCUUUUAUGUAAGCCA ..........((((....(((((((.((((....))))..............-..((..((...-((((((..(....)..))))))..))..))....)))))))((......)))))) ( -30.40) >DroYak_CAF1 33668 118 + 1 AGAACAGUAUUGGCAUUCGGCCAUAUGCCAUUUCUGGCCAUACUUAUAUCAA-ACAGGAGCAGC-AGCCGCAUCAGUUGGAGCGGUUAAGUGACUAAUUUAUGGCCUUUUAUGUAAGCCA ..........((((....(((((((.((((....))))..............-..((..((...-((((((..(....)..))))))..))..))....)))))))((......)))))) ( -30.40) >DroAna_CAF1 89753 118 + 1 AGAACUGUAUUGGCAUUCGGCCAUAUGCCAUUUCUGGCCAUACUUAUAUCAAAACAGGAGCA-C-GGCCGCAUCAGUUCGAGCGGUUAAGUGACUAAUUUAUGGCCUUUUGUGUAAGCCA ..........((((....(((.....)))......(((((((......((......))((((-(-.(((((.((.....)))))))...))).))....)))))))..........)))) ( -31.70) >consensus AGAACUGUAUUGGCAUUCGGCCAUAUGCCAUUUCUGGCCAUACUUAUAUCAA_ACAGGAGCAGC_AGCCGCAUCAGUUGGAGCGGUUAAGUGACUAAUUUAUGGCCUUUUAUGUAAGCCA ..........((((....(((((((.((((....)))).................((..((....((((((.((....)).))))))..))..))....)))))))((......)))))) (-29.88 = -29.77 + -0.11)


| Location | 4,881,649 – 4,881,767 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.75 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -30.93 |
| Energy contribution | -30.79 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4881649 118 - 22224390 UGGCUUACAUAAAAGGCCAUAAAUUAGUCACUUAACCGCUUCAACUGAUGCGGCU-GCUGCUCCUGU-UUGAUAUAAGUAUGGCCAGAAAUGGCAUAUGGCCGAAUGCCAAUACAGUUCU ((((..........(((((((........(((((.((((.((....)).))))..-...((....))-......)))))...((((....)))).)))))))....)))).......... ( -32.74) >DroSec_CAF1 32940 119 - 1 UGGCUUACAUAAAAGGCCAUAAAUUAGUCACUUAACCGCUCCAACUGAUGCGGCUGGCUGCUCCUGU-UUGAUAUAAGUAUGGCCAGAAAUGGCAUAUGGCCGAAUGCCAAUACAGUUCU ((((..........(((((((...((((((.....((((((.....)).)))).)))))).......-..............((((....)))).)))))))....)))).......... ( -33.24) >DroSim_CAF1 34123 118 - 1 UGGCUUACAUAAAAGGCCAUAAAUUAGUCACUUAACCGCUCCAACUGAUGCGGCU-GCUGCUCCUGU-UUGAUAUAAGUAUGGCCAGAAAUGGCAUAUGGCCGAAUGCCAAUACAGUUCU ((((..........(((((((........(((((.((((((.....)).))))..-...((....))-......)))))...((((....)))).)))))))....)))).......... ( -31.14) >DroEre_CAF1 35814 118 - 1 UGGCUUACAUAAAAGGCCAUAAAUUAGUCACUUAACCGCUCCAACUGAUGCGGCU-GCUGCUCCUGU-UUGAUAUAAGUAUGGCCAGAAAUGGCAUAUGGCCGAAUGCCAAUACAGUUCU ((((..........(((((((........(((((.((((((.....)).))))..-...((....))-......)))))...((((....)))).)))))))....)))).......... ( -31.14) >DroYak_CAF1 33668 118 - 1 UGGCUUACAUAAAAGGCCAUAAAUUAGUCACUUAACCGCUCCAACUGAUGCGGCU-GCUGCUCCUGU-UUGAUAUAAGUAUGGCCAGAAAUGGCAUAUGGCCGAAUGCCAAUACUGUUCU ((((..........(((((((........(((((.((((((.....)).))))..-...((....))-......)))))...((((....)))).)))))))....)))).......... ( -31.14) >DroAna_CAF1 89753 118 - 1 UGGCUUACACAAAAGGCCAUAAAUUAGUCACUUAACCGCUCGAACUGAUGCGGCC-G-UGCUCCUGUUUUGAUAUAAGUAUGGCCAGAAAUGGCAUAUGGCCGAAUGCCAAUACAGUUCU ((((((.......))))))......................((((((....((((-(-((((..(((....)))..))))))))).....((((((........))))))...)))))). ( -37.60) >consensus UGGCUUACAUAAAAGGCCAUAAAUUAGUCACUUAACCGCUCCAACUGAUGCGGCU_GCUGCUCCUGU_UUGAUAUAAGUAUGGCCAGAAAUGGCAUAUGGCCGAAUGCCAAUACAGUUCU ((((..........(((((((........(((((.((((((.....)).))))......((....)).......)))))...((((....)))).)))))))....)))).......... (-30.93 = -30.79 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:26 2006