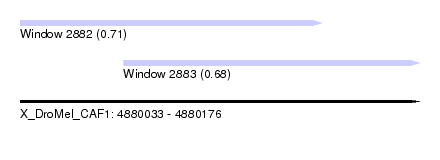
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,880,033 – 4,880,176 |
| Length | 143 |
| Max. P | 0.706457 |

| Location | 4,880,033 – 4,880,141 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.73 |
| Mean single sequence MFE | -24.08 |
| Consensus MFE | -17.01 |
| Energy contribution | -17.84 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.706457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
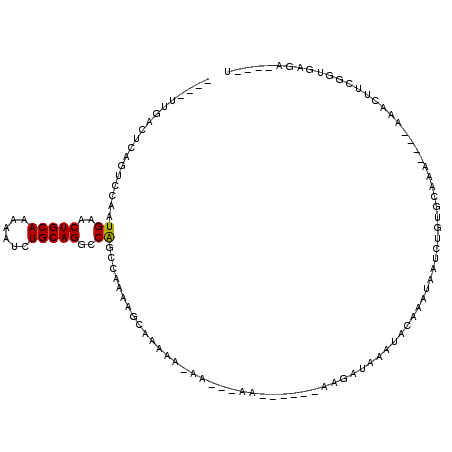
>X_DroMel_CAF1 4880033 108 + 22224390 UCAUUCACUUAGUCAGCAGACACUGGCGACUUGGUUU-------UUGACUCAGUCCAAUGAACUGCAAAAAUCUGCAGGCCAGCCAAAAGCAAAAAAAAAAAAA-----GAAAAUAAACA ...(((.....(((....))).(((((((((.((((.-------..)))).)))).......(((((......)))))))))).....................-----)))........ ( -24.60) >DroSec_CAF1 31313 104 + 1 UCAUUCACUUAGUCAGCAGACACUGGCGACUUGGUUU-------UUGACUCAGUCCAAUGAACUGCAAAAAUCUGCAGGCCAGCCAAAAGCAAAAA-AA---AA-----CAAGAUAAAUA .......(((.(((....))).(((((((((.((((.-------..)))).)))).......(((((......)))))))))).............-..---..-----.)))....... ( -23.90) >DroSim_CAF1 32566 105 + 1 UCAUUCACUUAGUCAGCAGACACUGGCGACUUGGUUU-------UUGACUCAGUCCAAUGAACUGCAAAAAUCUGCAGGCCAGCCAAAAGCAAAAAAAA---AA-----CAAGAUAAAUA .......(((.(((....))).(((((((((.((((.-------..)))).)))).......(((((......))))))))))................---..-----.)))....... ( -23.90) >DroEre_CAF1 34227 97 + 1 UCAUUCACUUAGUCAGCAGACACAGGCGACUUGGAUU-------UUGACUCAGUCCAAUGAACUGCAAAAAUCUGCAGGCCAGCCAAAAGAAAAA----------------AGAUAAGUA ......((((((((....)))...(((...(((((((-------.......)))))))((..(((((......)))))..)))))..........----------------...))))). ( -24.70) >DroYak_CAF1 32014 97 + 1 UCAUUCACUUAGUCAGCAGACACUGGCGACUUGGAUU-------UUGACUCAGUCCAAUGAACUGCAAAAAUCUGCAGGCCAGCCAAAAGGAAAA----------------AGAUAAAUA ...(((.(((.(((....))).(((((...(((((((-------.......)))))))....(((((......))))))))))....))))))..----------------......... ( -25.30) >DroAna_CAF1 84521 114 + 1 UCACUCACUUAGUCAGCAGACA--AGCGAAUUGGAUUUUUUUUUUCGACUCAGUCCAAUGAACUGCAAAAAUCUGCAGGCCGACCAAAAAAAUCAA-AA---AACCUCAGACACUAAAUA ...........(((.((.....--.))......(((((((((..(((..(((......))).(((((......)))))..)))..)))))))))..-..---.......)))........ ( -22.10) >consensus UCAUUCACUUAGUCAGCAGACACUGGCGACUUGGAUU_______UUGACUCAGUCCAAUGAACUGCAAAAAUCUGCAGGCCAGCCAAAAGCAAAAA_AA___AA______AAGAUAAAUA ...........(((....))).(((((((((.((..............)).)))).......(((((......))))))))))..................................... (-17.01 = -17.84 + 0.83)



| Location | 4,880,070 – 4,880,176 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.84 |
| Mean single sequence MFE | -15.50 |
| Consensus MFE | -8.32 |
| Energy contribution | -8.18 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4880070 106 + 22224390 ----UUGACUCAGUCCAAUGAACUGCAAAAAUCUGCAGGCCAGCCAAAAGCAAAAAAAAAAAAA-----GAAAAUAAACACAAAUAAUCUGUGCAAAAA-AAAACUUCGGUGAUA----U ----(((.(.(((.....((..(((((......)))))..))((.....)).............-----...................))).))))...-...((....))....----. ( -14.40) >DroSec_CAF1 31350 99 + 1 ----UUGACUCAGUCCAAUGAACUGCAAAAAUCUGCAGGCCAGCCAAAAGCAAAAA-AA---AA-----CAAGAUAAAUACAAAUAAUCUGUGCAAA----AAACUUCUGUGAGA----U ----....((((......((..(((((......)))))..)).......(((....-..---..-----..((((...........)))).)))...----.........)))).----. ( -14.50) >DroSim_CAF1 32603 100 + 1 ----UUGACUCAGUCCAAUGAACUGCAAAAAUCUGCAGGCCAGCCAAAAGCAAAAAAAA---AA-----CAAGAUAAAUACAAAUAAUCUGUGCAAA----AAACUUCGGUGAGA----U ----....((((..((..((..(((((......)))))..)).......(((.......---..-----..((((...........)))).)))...----.......)))))).----. ( -15.90) >DroEre_CAF1 34264 91 + 1 ----UUGACUCAGUCCAAUGAACUGCAAAAAUCUGCAGGCCAGCCAAAAGAAAAA----------------AGAUAAGUAUAAAUAAACUGUGCAAAA----AACAUUGGUGAAA----- ----.....(((..((((((..(((((......))))).................----------------......(((((.......)))))....----..)))))))))..----- ( -17.10) >DroYak_CAF1 32051 100 + 1 ----UUGACUCAGUCCAAUGAACUGCAAAAAUCUGCAGGCCAGCCAAAAGGAAAA----------------AGAUAAAUAAAAAUAAUCUGUGCAAAAAAAAAACUUUGGAGAAAACCUC ----.........(((((.(..(((((......)))))(((((((....))....----------------.................))).))..........).)))))......... ( -13.99) >DroAna_CAF1 84559 106 + 1 UUUUUCGACUCAGUCCAAUGAACUGCAAAAAUCUGCAGGCCGACCAAAAAAAUCAA-AA---AACCUCAGACACUAAAUAAAAACAAUUUGUU--AA----GAGUUUCGGUUAGA----U ....(((..(((......))).(((((......)))))..))).............-..---((((..((((.((.((((((.....))))))--.)----).)))).))))...----. ( -17.10) >consensus ____UUGACUCAGUCCAAUGAACUGCAAAAAUCUGCAGGCCAGCCAAAAGCAAAAA_AA___AA______AAGAUAAAUACAAAUAAUCUGUGCAAA____AAACUUCGGUGAGA____U ..................((..(((((......)))))..)).............................................................................. ( -8.32 = -8.18 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:20 2006