
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,872,375 – 4,872,484 |
| Length | 109 |
| Max. P | 0.571260 |

| Location | 4,872,375 – 4,872,484 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.41 |
| Mean single sequence MFE | -22.86 |
| Consensus MFE | -13.74 |
| Energy contribution | -13.97 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
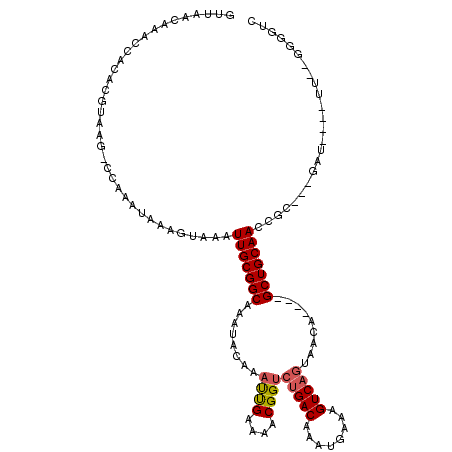
>X_DroMel_CAF1 4872375 109 + 22224390 GUUAACAAACCACACGUAAG-CCAAAUAAAGUAAAUUGCGGCAAAUACAAAUUGAAAACGGUCUGACAAAUGAAAGUCAGUAACA----GCUGCAACCGCUGCGAU----UU--GGGGAC .........((...(....)-((((((..(((...(((((((........((((....))))(((((........))))).....----)))))))..)))...))----))--)))).. ( -25.40) >DroSec_CAF1 23879 112 + 1 GUUAACAAACCACACGUAAG-CCAAAUAAAGUAAAUUGCGGCAAAUACAAAUUGAAAACGGUCUGACAAAUGAAAGUCAGUAACAACA-GCUGCAACCGCUGCGAU----UU--GGGAUC ....................-((((((..(((...(((((((........((((....))))(((((........)))))........-)))))))..)))...))----))--)).... ( -23.90) >DroSim_CAF1 24558 114 + 1 GGAAACAAACCUCUUUUUUUCCCCACCACCACAAAUUGCGGCCCCCCCCACCCGAAAACGGUCCGACCAACGAAAGUCAAUAACAACAGGCUGCAACCGCUGCCCA----UU--GGGAUC (....).............((((............((((((((........(((....)))...(((........)))..........))))))))..........----..--)))).. ( -23.75) >DroEre_CAF1 26818 106 + 1 GUUAACAAACCACACGUAAG-CCAAAUAAAGUAAAUUGCGGCAAAUACAAAUUGAAAACGGUCUGACAAAUGAAAGUCAGUAACA----GCUGCAACCGC---GAU----UU--GGGGUC ........(((...(....)-((((((...((...(((((((........((((....))))(((((........))))).....----)))))))..))---.))----))--))))). ( -24.10) >DroYak_CAF1 24134 110 + 1 GUUAACAAACUACACGUAAG-CCAAAUAAAGUAAAUUGCGGCAAAUACAAAUUGAAAACGGUCUGACAAAUGAAAGUCAGUAACA----GCUGCAACCGC---GAUUCGUUU--GGGGUC ....................-(((((..((.....(((((((........((((....))))(((((........))))).....----)))))))....---..))..)))--)).... ( -21.00) >DroAna_CAF1 77257 111 + 1 GUUAACAAGCCACACGUAAG-CCAAAUAAAGUAAAUUGCGGCAAAUACAAAUUGAAAACGGUCUGACAAAUGAAAGUCAGCAACA----GCUGCAACAAA---GUUAAA-AAGAGAUGGC ........(((((.(....)-..............(((((((........((((....))))(((((........))))).....----)))))))....---......-....).)))) ( -19.00) >consensus GUUAACAAACCACACGUAAG_CCAAAUAAAGUAAAUUGCGGCAAAUACAAAUUGAAAACGGUCUGACAAAUGAAAGUCAGUAACA____GCUGCAACCGC___GAU____UU__GGGGUC ...................................(((((((........((((....))))(((((........))))).........)))))))........................ (-13.74 = -13.97 + 0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:15 2006