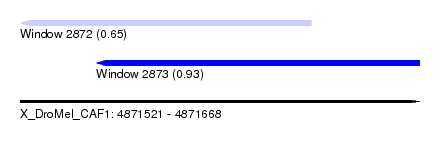
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,871,521 – 4,871,668 |
| Length | 147 |
| Max. P | 0.932273 |

| Location | 4,871,521 – 4,871,628 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.35 |
| Mean single sequence MFE | -22.82 |
| Consensus MFE | -15.10 |
| Energy contribution | -15.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
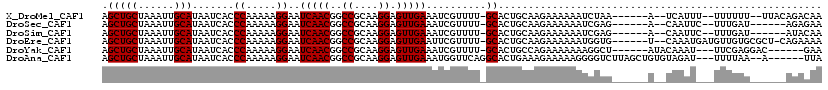
>X_DroMel_CAF1 4871521 107 - 22224390 AGCUGCUAAAUUGCAUAAUCACCCAAAAAGGAAUCAACGGCCGCAAGGAGUUGAAAUCGUUUU-GCACUGCAAGAAAAAAUCUAA------A--UCAUUU--UUUUUU--UUACAGACAA .(((((......)))..........((((.((.(((((..((....)).)))))..)).))))-)).(((.(((((((((.....------.--.....)--))))))--)).))).... ( -23.50) >DroSec_CAF1 23023 103 - 1 AGCUGCUAAAUUGCAUAAUCACCCAAAAAGGAAUCAACGGCCGCAAGGAGUUGAAAUCGUUUU-GCACUGCAAGAAAAAAUCGAG------A--CAAUUC--UUUGAU------AGAGAA .(((((......))........((.....)).......)))..((((((((((...(((((((-((...))))))......))).------.--))))))--))))..------...... ( -19.70) >DroSim_CAF1 23712 103 - 1 AGCUGCUAAAUUGCAUAAUCACCCAAAAAGGAAUCAACGGCCGCAAGGAGUUGAAAUCGUUUU-GCACUGCAAGAAAAAAUCGAG------A--CAAUUC--UUUGAU------AUACAA .(((((......))........((.....)).......)))..((((((((((...(((((((-((...))))))......))).------.--))))))--))))..------...... ( -19.70) >DroEre_CAF1 25901 110 - 1 AGCUGCUAAAUUGCAUAAUCACCCAAAAAGGAAUCAACGGCCGCAAGGAGUUGAAUUCGUUUU-GCACUGCAAGAAAAAAUGGUG------U--CAAAUGAUGUUGUGCGCU-CAGAAAA ..(((......((((((((((((......(((.(((((..((....)).))))).))).((((-((...))))))......))))------(--(....)).))))))))..-))).... ( -26.90) >DroYak_CAF1 23215 104 - 1 AGCUGCUAAAUUGCAUAAUCACCCAAAAAGGAAUCAACGGCCGCAAGGAGUUGAAAUCGUUUU-GCACUGCCAGAAAAAAAGGCU------AUACAAAU---UUCGAGGAC------GAA ...(((......)))...((..((.........(((((..((....)).)))))..(((.(((-(....(((.........))).------...)))).---..)))))..------)). ( -23.00) >DroAna_CAF1 76224 109 - 1 AGCUGCUAAAUUGCAUAAUCACCCAAAAAGGAAUCAACGGCCGCAAGGAGUUGAAAUGGUUCAGGCACUGAAAGAAAAAGGGGUCUUAGCUGUGUAGAU---UUUUAA--A------UUA ((((((......))......((((.........(((((..((....)).))))).....(((((...)))))........))))...))))........---......--.------... ( -24.10) >consensus AGCUGCUAAAUUGCAUAAUCACCCAAAAAGGAAUCAACGGCCGCAAGGAGUUGAAAUCGUUUU_GCACUGCAAGAAAAAAUCGAG______A__CAAAU___UUUGAU______AGACAA .(((((......))).......((.....))..(((((..((....)).)))))..........))...................................................... (-15.10 = -15.10 + -0.00)

| Location | 4,871,549 – 4,871,668 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.92 |
| Mean single sequence MFE | -28.77 |
| Consensus MFE | -24.34 |
| Energy contribution | -25.03 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4871549 119 - 22224390 GUGGUGCAGGCCAACAACGCCCCACAAACAAUCACUUACAAGCUGCUAAAUUGCAUAAUCACCCAAAAAGGAAUCAACGGCCGCAAGGAGUUGAAAUCGUUUU-GCACUGCAAGAAAAAA (..((((((((.......)))....((((..............(((......))).......((.....))..(((((..((....)).)))))....)))))-))))..)......... ( -29.00) >DroSec_CAF1 23047 119 - 1 GUGGUGCAGGCCAACAACGCCCCACAAACAAUCACUUACAAGCUGCUAAAUUGCAUAAUCACCCAAAAAGGAAUCAACGGCCGCAAGGAGUUGAAAUCGUUUU-GCACUGCAAGAAAAAA (..((((((((.......)))....((((..............(((......))).......((.....))..(((((..((....)).)))))....)))))-))))..)......... ( -29.00) >DroSim_CAF1 23736 119 - 1 GUGGUGCAGGCCAACAACGCCCCACAAACAAUCACUUACAAGCUGCUAAAUUGCAUAAUCACCCAAAAAGGAAUCAACGGCCGCAAGGAGUUGAAAUCGUUUU-GCACUGCAAGAAAAAA (..((((((((.......)))....((((..............(((......))).......((.....))..(((((..((....)).)))))....)))))-))))..)......... ( -29.00) >DroEre_CAF1 25932 119 - 1 GUGGUGCCGGCCAACAACGCCCCACAAACAAUCACUUACAAGCUGCUAAAUUGCAUAAUCACCCAAAAAGGAAUCAACGGCCGCAAGGAGUUGAAUUCGUUUU-GCACUGCAAGAAAAAA (..((((.(((.......)))......................(((......)))..........((((.((((((((..((....)).)))).)))).))))-))))..)......... ( -29.60) >DroYak_CAF1 23240 119 - 1 GUGGUGCAGGCCAACAACGCCCCACAAACAAUCACUUACAAGCUGCUAAAUUGCAUAAUCACCCAAAAAGGAAUCAACGGCCGCAAGGAGUUGAAAUCGUUUU-GCACUGCCAGAAAAAA (..((((((((.......)))....((((..............(((......))).......((.....))..(((((..((....)).)))))....)))))-))))..)......... ( -28.10) >DroAna_CAF1 76253 118 - 1 GCCGUGCC--CGAACAACGCCCCACAAACAAUCACUUACAAGCUGCUAAAUUGCAUAAUCACCCAAAAAGGAAUCAACGGCCGCAAGGAGUUGAAAUGGUUCAGGCACUGAAAGAAAAAG ...(((((--.((((............................(((......))).......((.....))..(((((..((....)).)))))....)))).)))))............ ( -27.90) >consensus GUGGUGCAGGCCAACAACGCCCCACAAACAAUCACUUACAAGCUGCUAAAUUGCAUAAUCACCCAAAAAGGAAUCAACGGCCGCAAGGAGUUGAAAUCGUUUU_GCACUGCAAGAAAAAA (((((((.(((.......)))....((((..............(((......))).......((.....))..(((((..((....)).)))))....))))..)))))))......... (-24.34 = -25.03 + 0.70)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:11 2006