| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,871,045 – 4,871,153 |
| Length | 108 |
| Max. P | 0.552974 |

| Location | 4,871,045 – 4,871,153 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.60 |
| Mean single sequence MFE | -27.43 |
| Consensus MFE | -18.13 |
| Energy contribution | -17.97 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4871045 108 + 22224390 ----AA------AAGAAUAACUUUGGCCAAGAAGUUUUUGACUCGUUGUUGGCCU-UUGUUUGCUUUGCAAAUUGCGCUCAAGUGCGCCGC-UAAGAUAAUCAGUGCAAAUUCUGGCCAA ----..------..........(((((((.(((...(((((((.(((((((((..-..((((((...)))))).((((......)))).))-)..)))))).))).)))))))))))))) ( -30.90) >DroSec_CAF1 22572 111 + 1 ----AAAAGAAAAAGAAUAACU-UGGCCAAGA-GAUUUUGAUUCGUU-UUGGCCU-UUGUUUGCUUUGCAAAUUGCGCUCAAGUGCGCCGC-CGAGAUAAUCAGUGCAAAUUCUGGCCAA ----..................-((((((..(-....((((((.(((-(((((..-..((((((...)))))).((((......)))).))-))))))))))))......)..)))))). ( -34.70) >DroSim_CAF1 23255 110 + 1 ----AAAA-AGAAAGAAUAACU-UGGCCAAGA-GAUUUUGACUCGUU-UUGGCCU-UUGUUUGCUUUGCAAAUUGCGCUCAAGUGCGCCGG-CGAGAUAAUCAGUGCAAAUUCUGGCCAA ----..((-((...((((((..-.((((((((-((.......)).))-)))))).-)))))).)))).......((((......)))).((-(.(((..............))).))).. ( -31.04) >DroEre_CAF1 25449 112 + 1 --AAAAAA-AAAAAGAAUAACU-UAGCCAAGA-GAUUUUGACUCCUU-UUGGUCU-UUGUUUGCUUUGCAAAUUGCGCUCAAGUGCGCCUC-UGAGAUAAUCAGUGCAAAUUCUGGCCAA --......-....((((((((.-.((((((((-(..........)))-)))).))-..)))....(((((....((((......))))..(-(((.....))))))))))))))...... ( -26.80) >DroYak_CAF1 22773 114 + 1 AUAAAAAA-AAAAAGAAUAACU-UAGCCAAGA-GAUUUUGACUCCUU-UUGGUCU-UUGUUUGCUUUGCAAAUUGCGCUCAAGUGCGCCUC-CGAGAUAAUCAGUGCAAAUUCUGGCCAA ........-....((((((((.-.((((((((-(..........)))-)))).))-..)))....(((((....((((......))))...-.((.....))..))))))))))...... ( -23.90) >DroAna_CAF1 75525 91 + 1 -------------------------UCUCAAA-GACUCAGACUCCUUUUUGGCCUUUUGUUUGCUUUGCAAAUUGCACUCAAGUGCACCUCCCCAGAUAAUCAGUGCAAAUUCUGGC--- -------------------------...((((-(...((((..((.....))...))))....))))).....(((((....))))).....(((((..............))))).--- ( -17.24) >consensus ____AAAA_AAAAAGAAUAACU_UGGCCAAGA_GAUUUUGACUCCUU_UUGGCCU_UUGUUUGCUUUGCAAAUUGCGCUCAAGUGCGCCGC_CGAGAUAAUCAGUGCAAAUUCUGGCCAA .................................................(((((....((((((.(((.....(((((....))))).(......).....))).))))))...))))). (-18.13 = -17.97 + -0.16)

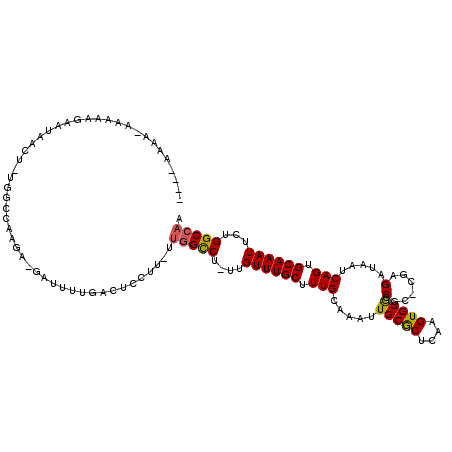

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:09 2006