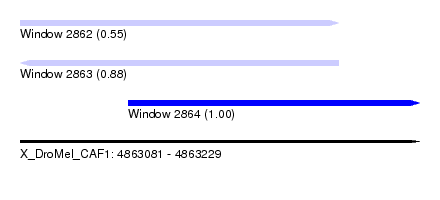
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,863,081 – 4,863,229 |
| Length | 148 |
| Max. P | 0.996225 |

| Location | 4,863,081 – 4,863,199 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 95.85 |
| Mean single sequence MFE | -32.68 |
| Consensus MFE | -27.43 |
| Energy contribution | -27.71 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4863081 118 + 22224390 GGCCCGGUUGGUUGCUGCUCCAAUUGGCCAACUGAUGCUAACGGCCAACUGUUCGAUCGCUGCACAAAUUAUAAUUUGCCAAGUAUUCGCAUUUUAGCGGCAACUUUGUCGCCAGAAU (((......((((((((((.((.((((((.............)))))).))..(((..(((...(((((....)))))...)))..)))......)))))))))).....)))..... ( -35.52) >DroSec_CAF1 14859 114 + 1 GGCCCGGU----UGCUGCUCCAAUUGGCCAACUGAUGCUAACGGCCAACUGUUCGAUCGCUGCACAAAUUAUAAUUUGCCAAGUAUUCGCAUUUUAGCGGCAACUUUGUCGCCAGAAU (((..(((----(((((((.((.((((((.............)))))).))..(((..(((...(((((....)))))...)))..)))......)))))))))).....)))..... ( -34.22) >DroSim_CAF1 14845 114 + 1 GGCCCGGU----UGCUGCUCCAAUUGGCCAACUGAUGCUAACGGCCAACUGUUCGAUCGCUGCACAAAUUAUAAUUUGCCAAGUAUUCGCAUUUUAGCGGCAACUUUGUCGCCAGAAU (((..(((----(((((((.((.((((((.............)))))).))..(((..(((...(((((....)))))...)))..)))......)))))))))).....)))..... ( -34.22) >DroEre_CAF1 17889 113 + 1 GGUCCGU-----UGCUGCUCCAAUUGGCCAACUGAUGCUAACGGCCAACUGUUCGAUCGCCGCACAAAUUAUAAUAUGCCAAGUAUUCGCAUUUUAGCGGCAACUUUGUCGCCAGAAU (((..((-----((((((.....((((((.............)))))).(((.((.....)).))).....(((.((((.........)))).)))))))))))......)))..... ( -26.72) >DroYak_CAF1 15062 114 + 1 GGCCCGUU----UGCUGCUCCAAUUGGCCAACUGAUGCUAACGGCCAACUGUUUGAUGGCCGCACAAAUUAUAAUUUGCCAAGUAUUCGCAUUUUAGCGGCAACUUUGUCGCCAGAAU ((((...(----((......)))..))))..(((((((...((((((.(.....).))))))..(((((....)))))..........))))....((((((....)))))))))... ( -32.70) >consensus GGCCCGGU____UGCUGCUCCAAUUGGCCAACUGAUGCUAACGGCCAACUGUUCGAUCGCUGCACAAAUUAUAAUUUGCCAAGUAUUCGCAUUUUAGCGGCAACUUUGUCGCCAGAAU (((.........(((((((.((.((((((.............)))))).))..(((..(((...(((((....)))))...)))..)))......)))))))........)))..... (-27.43 = -27.71 + 0.28)

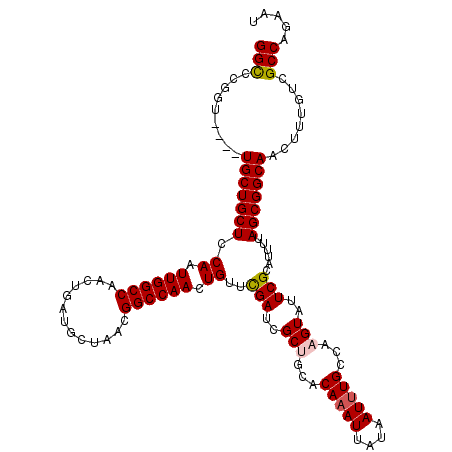
| Location | 4,863,081 – 4,863,199 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 95.85 |
| Mean single sequence MFE | -34.68 |
| Consensus MFE | -32.18 |
| Energy contribution | -32.58 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4863081 118 - 22224390 AUUCUGGCGACAAAGUUGCCGCUAAAAUGCGAAUACUUGGCAAAUUAUAAUUUGUGCAGCGAUCGAACAGUUGGCCGUUAGCAUCAGUUGGCCAAUUGGAGCAGCAACCAACCGGGCC ((((.((((((...))))))((......)))))).((((((((((....)))))(((.((.......((((((((((...........))))))))))..)).))).....))))).. ( -34.70) >DroSec_CAF1 14859 114 - 1 AUUCUGGCGACAAAGUUGCCGCUAAAAUGCGAAUACUUGGCAAAUUAUAAUUUGUGCAGCGAUCGAACAGUUGGCCGUUAGCAUCAGUUGGCCAAUUGGAGCAGCA----ACCGGGCC ((((.((((((...))))))((......)))))).((((((((((....)))))(((.((.......((((((((((...........))))))))))..)).)))----.))))).. ( -34.70) >DroSim_CAF1 14845 114 - 1 AUUCUGGCGACAAAGUUGCCGCUAAAAUGCGAAUACUUGGCAAAUUAUAAUUUGUGCAGCGAUCGAACAGUUGGCCGUUAGCAUCAGUUGGCCAAUUGGAGCAGCA----ACCGGGCC ((((.((((((...))))))((......)))))).((((((((((....)))))(((.((.......((((((((((...........))))))))))..)).)))----.))))).. ( -34.70) >DroEre_CAF1 17889 113 - 1 AUUCUGGCGACAAAGUUGCCGCUAAAAUGCGAAUACUUGGCAUAUUAUAAUUUGUGCGGCGAUCGAACAGUUGGCCGUUAGCAUCAGUUGGCCAAUUGGAGCAGCA-----ACGGACC ..((((((..(...((((((((......)).........(((((........)))))))))))....((((((((((...........))))))))))..)..)).-----.)))).. ( -34.40) >DroYak_CAF1 15062 114 - 1 AUUCUGGCGACAAAGUUGCCGCUAAAAUGCGAAUACUUGGCAAAUUAUAAUUUGUGCGGCCAUCAAACAGUUGGCCGUUAGCAUCAGUUGGCCAAUUGGAGCAGCA----AACGGGCC .....((((((...))))))(((....(((.........((((((....))))))(((((((.(.....).)))))))..((..(((((....)))))..)).)))----....))). ( -34.90) >consensus AUUCUGGCGACAAAGUUGCCGCUAAAAUGCGAAUACUUGGCAAAUUAUAAUUUGUGCAGCGAUCGAACAGUUGGCCGUUAGCAUCAGUUGGCCAAUUGGAGCAGCA____ACCGGGCC ((((.((((((...))))))((......)))))).((((((((((....))))))((.((.......((((((((((...........))))))))))..)).)).......)))).. (-32.18 = -32.58 + 0.40)



| Location | 4,863,121 – 4,863,229 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 92.84 |
| Mean single sequence MFE | -33.68 |
| Consensus MFE | -27.56 |
| Energy contribution | -28.12 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4863121 108 + 22224390 ACGGCCAACUGUUCGAUCGCUGCACAAAUUAUAAUUUGCCAAGUAUUCGCAUUUUAGCGGCAACUUUGUCGCCAGAAUGCG-GAUUGGAAUUACCACAA-----CCAGCGGUUU ((((....))))..((((((((..(((((....)))))(((...(((((((((((.((((((....)))))).))))))))-))))))...........-----.)))))))). ( -35.30) >DroSec_CAF1 14895 113 + 1 ACGGCCAACUGUUCGAUCGCUGCACAAAUUAUAAUUUGCCAAGUAUUCGCAUUUUAGCGGCAACUUUGUCGCCAGAAUGCG-GAUUGGAAUUACCACAACACAACCAGCAGUUU .((..(....)..))...(((((.(((((....)))))(((...(((((((((((.((((((....)))))).))))))))-))))))...................))))).. ( -33.30) >DroSim_CAF1 14881 114 + 1 ACGGCCAACUGUUCGAUCGCUGCACAAAUUAUAAUUUGCCAAGUAUUCGCAUUUUAGCGGCAACUUUGUCGCCAGAAUGCGGGAUUGGAAUUACCACAACACAGCCAGCAGUUU ......(((((((.....((((..(((((....)))))((((...((((((((((.((((((....)))))).)))))))))).)))).............)))).))))))). ( -34.00) >DroEre_CAF1 17924 107 + 1 ACGGCCAACUGUUCGAUCGCCGCACAAAUUAUAAUAUGCCAAGUAUUCGCAUUUUAGCGGCAACUUUGUCGCCAGAAUGCG-GAUUGGAAUUACCACAA-----CUAGGAGU-U .((((.............))))................((.((((((((((((((.((((((....)))))).))))))))-)))(((.....)))..)-----)).))...-. ( -29.92) >DroYak_CAF1 15098 107 + 1 ACGGCCAACUGUUUGAUGGCCGCACAAAUUAUAAUUUGCCAAGUAUUCGCAUUUUAGCGGCAACUUUGUCGCCAGAAUGCG-GAUUGGAAUUGCCACAA-----CUAGCUGU-U ..(((((.(.....).)))))((((((((....)))))(((...(((((((((((.((((((....)))))).))))))))-))))))...))).....-----........-. ( -35.90) >consensus ACGGCCAACUGUUCGAUCGCUGCACAAAUUAUAAUUUGCCAAGUAUUCGCAUUUUAGCGGCAACUUUGUCGCCAGAAUGCG_GAUUGGAAUUACCACAA_____CCAGCAGUUU .((..(....)..))...(((((.(((((....)))))((((.....((((((((.((((((....)))))).))))))))...))))...................))))).. (-27.56 = -28.12 + 0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:18:03 2006