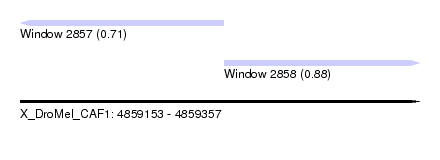
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,859,153 – 4,859,357 |
| Length | 204 |
| Max. P | 0.878373 |

| Location | 4,859,153 – 4,859,257 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 96.38 |
| Mean single sequence MFE | -19.82 |
| Consensus MFE | -18.90 |
| Energy contribution | -19.10 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4859153 104 - 22224390 UUGGUAGGAGUGUUCCGUUAAAAGCAAGAAACUACUCAUUAUUCCA-AACAUUUCACCCACUUUUGGUAUAUUACGACGCCGCUACUCCUCCCACUUU-UCUGUAU .(((.((((((((..((((........((......))..(((.(((-((.............))))).)))....))))..)).)))))).)))....-....... ( -18.92) >DroSec_CAF1 11017 105 - 1 UUGGUAGGAGUGUUCCGUUAAAAGCUAGAAACUACUCAUUAUUCCAAAACAUUUCACCCACUUUUGGUGUAUUACGACGCCGCUACUCCUCCCACUGU-UCUGUAU .(((.((((((((..((((........((......))..(((.((((((............)))))).)))....))))..)).)))))).)))....-....... ( -20.20) >DroSim_CAF1 11002 105 - 1 UUGGUAGGAGUGUUCCGUUAAAAGCUAGAAACUACUCAUUAUUCCAAAACAUUUCACCCACUUUUGGUGUAUUACGACGCCGCUACUCCUCCCACUGU-UCUGUAU .(((.((((((((..((((........((......))..(((.((((((............)))))).)))....))))..)).)))))).)))....-....... ( -20.20) >DroEre_CAF1 12490 104 - 1 CUGGUAGGAGUGUUCCGUUAAAAGCUAGAAACUACUCAUUAUUCCA-AACAUUUCACCCACUUUUGGUGUAUUACGACGCCGCUACUCCUCCCACUUU-UUUGUAU .(((.((((((((..((((........((......)).........-.......((((.......))))......))))..)).)))))).)))....-....... ( -19.80) >DroYak_CAF1 11225 105 - 1 UUGGUAGGAGUGUUCCGUUUAAAGCUAGAAACUACUCAUUAUUCCA-AACAUUUCACCCACUUUUGGUGUAUUACGACGCCGCUACUCCUCCCACUUUUUUUGUAU .(((.(((((((....((((........))))..............-..................(((((......)))))..))))))).)))............ ( -20.00) >consensus UUGGUAGGAGUGUUCCGUUAAAAGCUAGAAACUACUCAUUAUUCCA_AACAUUUCACCCACUUUUGGUGUAUUACGACGCCGCUACUCCUCCCACUUU_UCUGUAU .(((.((((((((..((((........((......)).................((((.......))))......))))..)).)))))).)))............ (-18.90 = -19.10 + 0.20)

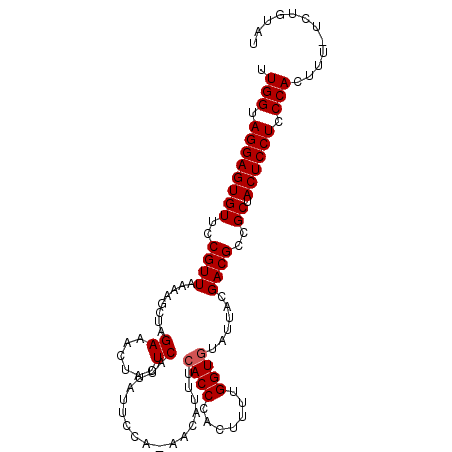

| Location | 4,859,257 – 4,859,357 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.80 |
| Mean single sequence MFE | -20.76 |
| Consensus MFE | -3.18 |
| Energy contribution | -4.34 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.15 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4859257 100 + 22224390 GCAGAACAAC-UGGAAGCAAUGUUCUGU-----UUCGGCUUAGGUUCGC-AAAAAACGAAGUUAAGCGUACUUAAAAAAUAUAA------A-----GAAAGA--AAACUGCAUGUUUUCG ((((((((..-((....)).))))))))-----....((((((.((((.-......)))).)))))).................------.-----....((--((((.....)))))). ( -26.50) >DroSec_CAF1 11122 102 + 1 GCAGAACAAC-UGCAAGCAAUGUUCUCUUU--UUUUGGCUUAGGUUCGC-AAAAAACUAAGUUAAGCAUACA-AAAAUAUAUGA------A-----GAAAGA--AAACUGCAUGUUUUCG ((((.....)-)))..(((..(((.(((((--(((((((((((......-......))))))))..((((..-......)))))------)-----))))))--.))))))......... ( -21.70) >DroSim_CAF1 11107 104 + 1 GCAGAACAAC-UGCAAGCAAUGUUCUCUUUUUUUUUGGCUUAGGUUCGC-AAAAAACUAAGUUAAUCAUACA-AAAAUAUAUAG------A-----GAAAGA--AAACUGCAUGUUUUCG ((((.....)-)))........((((((...((((((((((((......-......))))))........))-)))).....))------)-----))).((--((((.....)))))). ( -22.60) >DroEre_CAF1 12594 109 + 1 GCGGAACAACUAAAAAGCAAUGUUC--------UUUGGCCUAAGUUCGCAUAAAAACUAAGUUAGAGAUACA-AAAAUAUAAAGUUUGCUAAAACUGAAAGA--AAACUAAAUGUUUUAA .............((((((.(((((--------((((((...((((........))))..)))))))).)))-.........(((((....)))))......--........)))))).. ( -15.10) >DroYak_CAF1 11330 106 + 1 GCAGAACAAC-AAAAACCAGUGGAC--------UUUGGUUUAGAUUCGC-CAAAAACUAAGUAAAGGAUACA-AAAAUACAAAGUUUGCUAGAAUCUAAAUCUAUGCUUAAAUCUUA--- (((.......-..(((((((.....--------.)))))))((((((..-...(((((..(((.........-....)))..)))))....)))))).......)))..........--- ( -17.92) >consensus GCAGAACAAC_UGAAAGCAAUGUUCU_U_____UUUGGCUUAGGUUCGC_AAAAAACUAAGUUAAGCAUACA_AAAAUAUAUAG______A_____GAAAGA__AAACUGCAUGUUUUCG ...(((((............)))))........((((((((((.............))))))))))...................................................... ( -3.18 = -4.34 + 1.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:57 2006