
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,858,986 – 4,859,096 |
| Length | 110 |
| Max. P | 0.729458 |

| Location | 4,858,986 – 4,859,096 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.15 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -17.83 |
| Energy contribution | -19.33 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4858986 110 + 22224390 CAACGCCUCAUAUG----UUUU---U--CUUUUUUCGAAAUGGGAACGCUCUUGAUUAUUCCUUUGUUUGCCGCCUGCCUUCUCUAAAAGGGGGGCGUGGCACGAGUGGUCCACAUU-UG ..............----....---.--.........((((((((.(((((..((....)).......((((((..(((((((......))))))))))))).))))).))).))))-). ( -31.50) >DroSec_CAF1 10849 111 + 1 CAACGCCUCAUAUG----UUUU---UUUUUUCUUUCGAAAUGGGAACGCUCUUGAUUAUUCCUUUGUUUGCCGCCUGCCUUCUCUAAAAGG-GGGCGUGGCACGAGUGGUCCACAUU-UG ..............----....---............((((((((.(((((..((....)).......((((((..(((((((.....)))-)))))))))).))))).))).))))-). ( -31.30) >DroSim_CAF1 10830 115 + 1 CAACGCCUCAUAUGCUUCUUUU---UUUUUUCUUUCGAAAUGGGAACGCUCUUGAUUAUUCCUUUGUUUGCCGCCUGCCUUCUCUAAAAGG-GGGCGUGGCACGAGUGGUCCACAUU-UG ......................---............((((((((.(((((..((....)).......((((((..(((((((.....)))-)))))))))).))))).))).))))-). ( -31.30) >DroEre_CAF1 12338 101 + 1 CAACGCCUCAUAUG----A------------UUUUCGAAAUGGGAACGCUCUUGAUUAUUCCUUUGU-UGCCGCCUGCCUUCUCUAAAAGG-GGGCGUGGCUCGAGUGGUCCACAUU-UG ..............----.------------......((((((((.(((((..((....))......-.(((((..(((((((.....)))-)))))))))..))))).))).))))-). ( -30.20) >DroYak_CAF1 11074 100 + 1 GAACGCCUCAUGUU-----------------UUUGCGAAAUGGGAACGCUCUUGAUUAUUCCUUUGU-UGUCGCUAGCCUUCUCUAAAAGG-GGGCGUGGCACGAGUGGUCCACAUU-UG ...(((........-----------------...)))((((((((.(((((..(((.((......))-.)))(((((((((((.....)))-)))).))))..))))).))).))))-). ( -31.50) >DroAna_CAF1 63030 100 + 1 UAAUGCCUCAAUCG----AUUUAGUGUUCUUCUACUUAAAUUAGAACACUAUUGAUUAUUCCUUA---------------CCUCGGAAAAG-GGGCGGGUCUGGCAUGCUCUACAUUUUG ..(((((..(((((----(..(((((((((............)))))))))))))))....((..---------------((((......)-)))..))...)))))............. ( -27.80) >consensus CAACGCCUCAUAUG____UUUU___U___UUCUUUCGAAAUGGGAACGCUCUUGAUUAUUCCUUUGU_UGCCGCCUGCCUUCUCUAAAAGG_GGGCGUGGCACGAGUGGUCCACAUU_UG ......................................(((((((.(((((..((....))........(((((..((((.((......)).)))))))))..))))).))).))))... (-17.83 = -19.33 + 1.50)
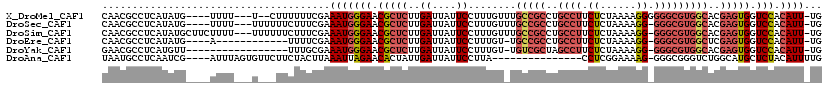

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:56 2006