
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,858,839 – 4,858,946 |
| Length | 107 |
| Max. P | 0.768304 |

| Location | 4,858,839 – 4,858,946 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.00 |
| Mean single sequence MFE | -26.93 |
| Consensus MFE | -17.52 |
| Energy contribution | -18.40 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
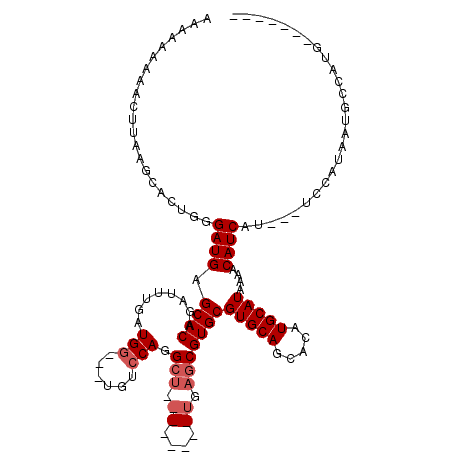
>X_DroMel_CAF1 4858839 107 - 22224390 AAAAAAAAACUUAAGCACUGGGAUGAGCACAGAUUUGAUGG---UGUCCAGGCUGAGCUGGCUGAGCGUGCGUGCAGCACAUGCAUAAAACAUCAU---UCCAUAAUGCCAUG------- ..............(((.((((((((((((........(((---...))).(((.((....)).)))))))(((((.....)))))......))))---))))...)))....------- ( -31.00) >DroSec_CAF1 10716 100 - 1 AAAAAAAAACUUAAGCACUGGGAUGAGCACAGAUUUGAUGG---UGUCCAGGCUGAGCUGGCUGAGCGUGCGUGCAUCACAUGCAUAAAACAUCAU---UCCAUGA-------------- ...............((.((((((((((((........(((---...))).(((.((....)).)))))))((((((...))))))......))))---)))))).-------------- ( -28.70) >DroSim_CAF1 10697 100 - 1 AAAAAAAAACUUAAGCACUGGGAUGAGCACAGAUUUGAUGG---UGUCCAGGCUGAGCUGGCUGAGCGUGCGUGCAGCACAUGCAUAAAACAUCAU---UCCAUAA-------------- ..................((((((((((((........(((---...))).(((.((....)).)))))))(((((.....)))))......))))---))))...-------------- ( -28.80) >DroEre_CAF1 12212 93 - 1 AAA-----ACUUAAGCGCUGGGAUGAGCACGGAUUUGAUGG---UGUCCAGGCU---------GAGCGUGCGUGCAGCACAUGCAUAAAACAUCAA---UCCAUAAUGCCAUG------- ...-----......(((((......)))..(((((.((((.---(((.((.(((---------(.((....)).))))...)).)))...))))))---))).....))....------- ( -24.30) >DroYak_CAF1 10943 98 - 1 AAAAAAUGACUUAAGCGCUGGGAUGAGCACAGAUUUGAUGG---UGUCCAGGCU---------GAGCGUGCGUGCAGCACAUGCAUAAAACAUCAU---UCCAUAAUGCCAUG------- ..............(((.((((((((((.(((.((((....---....))))))---------).))(((((((.....)))))))......))))---))))...)))....------- ( -26.20) >DroAna_CAF1 62905 107 - 1 --AAAAAAACAC-AACACAGAGAUGAGCACAAAUUUGAUGAUGAUGUGCAGG----------UGUGCGUGCGUGCAACACAUGCAUAAAACAUCAAAAGCCCAUAAUGCCAUGCAGAAUC --..........-....((...(((.((.....(((((((.(..((((((.(----------((((((....))).)))).))))))..)))))))).)).)))..))............ ( -22.60) >consensus AAAAAAAAACUUAAGCACUGGGAUGAGCACAGAUUUGAUGG___UGUCCAGGCU________UGAGCGUGCGUGCAGCACAUGCAUAAAACAUCAU___UCCAUAAUGCCAUG_______ .....................((((.((((........(((......))).(((.((....)).)))))))(((((.....)))))....)))).......................... (-17.52 = -18.40 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:55 2006