
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,857,729 – 4,857,838 |
| Length | 109 |
| Max. P | 0.989217 |

| Location | 4,857,729 – 4,857,821 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.47 |
| Mean single sequence MFE | -23.83 |
| Consensus MFE | -21.59 |
| Energy contribution | -21.78 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4857729 92 + 22224390 ----------------------------CAGCUGCUGUUGGCAAUUUAUAAACGAAAUAAAUUCAAUUGACUUGUUUGCCGCAGCAUUUACAGUUGGCAACAAUUGCAGCAAUGCCAUAA ----------------------------..((((((((.(((((.........(((.....)))...........)))))))))).....((((((....)))))).))).......... ( -23.45) >DroSec_CAF1 9661 92 + 1 ----------------------------CAGCUGCUGUUGGCAAUUUAUAAACGAAAUAAAUUCAAUUGACUUGUUUGCCGCAGCAUUUACAGUUGGCAACAAUUGCAGCAAUGCCAUAA ----------------------------..((((((((.(((((.........(((.....)))...........)))))))))).....((((((....)))))).))).......... ( -23.45) >DroSim_CAF1 9628 92 + 1 ----------------------------CAGCUGCUGUUGGCAAUUUAUAAACGAAAUAAAUUCAAUUGACUUGUUUGCCGCAGCAUUUACAGUUGGCAACAAUUGCAGCAAUGCCAUAA ----------------------------..((((((((.(((((.........(((.....)))...........)))))))))).....((((((....)))))).))).......... ( -23.45) >DroEre_CAF1 11145 92 + 1 ----------------------------CAGCUGCUGUUGGCAAUUUAUAAACGAAAUAAAUUCAAUUGACUUGUUUGCCGCAGCAUUUACAGUUGGCAACAAUUGCAGCAAUGCCAUAA ----------------------------..((((((((.(((((.........(((.....)))...........)))))))))).....((((((....)))))).))).......... ( -23.45) >DroYak_CAF1 9834 92 + 1 ----------------------------CAGCUGCUGUUGGCAAUUUAUAAACGAAAUAAAUUCAAUUGACUUGUUUGCCGCAGCAUUUACAGUUGGCAACAAUUGCAGCAAUGCCAUAA ----------------------------..((((((((.(((((.........(((.....)))...........)))))))))).....((((((....)))))).))).......... ( -23.45) >DroAna_CAF1 61366 120 + 1 CACCGUCGACUCCACACCGUCGAUUCCACUGAUUCUGCUGGCAAUUUAUAAACGAAAUAAAUUCAAUUGACUUGUUUGCCGCAGCAUUUACAGUUGGCAACAAUUGCAGCAAUGCCAUAA ....((((((........))))))....(((...((((.(((((.........(((.....)))...........)))))))))......((((((....)))))))))........... ( -25.75) >consensus ____________________________CAGCUGCUGUUGGCAAUUUAUAAACGAAAUAAAUUCAAUUGACUUGUUUGCCGCAGCAUUUACAGUUGGCAACAAUUGCAGCAAUGCCAUAA ..............................((((((((.(((((.........(((.....)))...........)))))))))).....((((((....)))))).))).......... (-21.59 = -21.78 + 0.20)

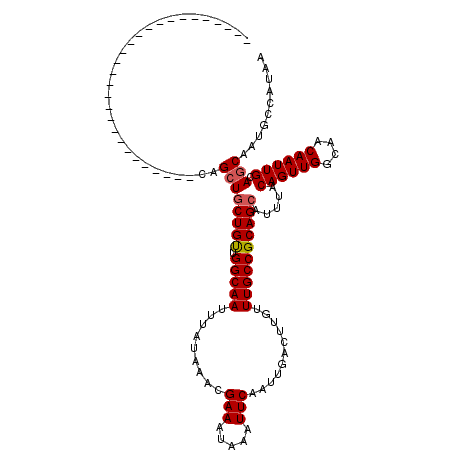
| Location | 4,857,729 – 4,857,821 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.47 |
| Mean single sequence MFE | -22.53 |
| Consensus MFE | -21.20 |
| Energy contribution | -21.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4857729 92 - 22224390 UUAUGGCAUUGCUGCAAUUGUUGCCAACUGUAAAUGCUGCGGCAAACAAGUCAAUUGAAUUUAUUUCGUUUAUAAAUUGCCAACAGCAGCUG---------------------------- ...((((.((((((((....((((.....))))....))))))))............(((((((.......)))))))))))..........---------------------------- ( -21.20) >DroSec_CAF1 9661 92 - 1 UUAUGGCAUUGCUGCAAUUGUUGCCAACUGUAAAUGCUGCGGCAAACAAGUCAAUUGAAUUUAUUUCGUUUAUAAAUUGCCAACAGCAGCUG---------------------------- ...((((.((((((((....((((.....))))....))))))))............(((((((.......)))))))))))..........---------------------------- ( -21.20) >DroSim_CAF1 9628 92 - 1 UUAUGGCAUUGCUGCAAUUGUUGCCAACUGUAAAUGCUGCGGCAAACAAGUCAAUUGAAUUUAUUUCGUUUAUAAAUUGCCAACAGCAGCUG---------------------------- ...((((.((((((((....((((.....))))....))))))))............(((((((.......)))))))))))..........---------------------------- ( -21.20) >DroEre_CAF1 11145 92 - 1 UUAUGGCAUUGCUGCAAUUGUUGCCAACUGUAAAUGCUGCGGCAAACAAGUCAAUUGAAUUUAUUUCGUUUAUAAAUUGCCAACAGCAGCUG---------------------------- ...((((.((((((((....((((.....))))....))))))))............(((((((.......)))))))))))..........---------------------------- ( -21.20) >DroYak_CAF1 9834 92 - 1 UUAUGGCAUUGCUGCAAUUGUUGCCAACUGUAAAUGCUGCGGCAAACAAGUCAAUUGAAUUUAUUUCGUUUAUAAAUUGCCAACAGCAGCUG---------------------------- ...((((.((((((((....((((.....))))....))))))))............(((((((.......)))))))))))..........---------------------------- ( -21.20) >DroAna_CAF1 61366 120 - 1 UUAUGGCAUUGCUGCAAUUGUUGCCAACUGUAAAUGCUGCGGCAAACAAGUCAAUUGAAUUUAUUUCGUUUAUAAAUUGCCAGCAGAAUCAGUGGAAUCGACGGUGUGGAGUCGACGGUG ...((((....(..((.(((((((((.(((......(((((((...(((.....)))(((((((.......)))))))))).))))...))))))...))))))))..).))))...... ( -29.20) >consensus UUAUGGCAUUGCUGCAAUUGUUGCCAACUGUAAAUGCUGCGGCAAACAAGUCAAUUGAAUUUAUUUCGUUUAUAAAUUGCCAACAGCAGCUG____________________________ ...((((.((((((((....((((.....))))....))))))))............(((((((.......)))))))))))...................................... (-21.20 = -21.20 + -0.00)

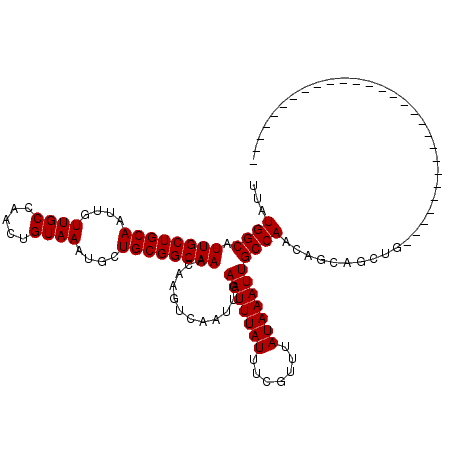

| Location | 4,857,741 – 4,857,838 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.04 |
| Mean single sequence MFE | -20.03 |
| Consensus MFE | -18.52 |
| Energy contribution | -18.52 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4857741 97 + 22224390 GCAAUUUAUAAACGAAAUAAAUUCAAUUGACUUGUUUGCCGCAGCAUUUACAGUUGGCAACAAUUGCAGCAAUGCCAUAAAUCGUUGGACAAGCAUU----------------------- (.(((((((.......))))))))........((((((.(.(((((((((.(((((....)))))(((....)))..))))).))))).))))))..----------------------- ( -20.20) >DroSec_CAF1 9673 97 + 1 GCAAUUUAUAAACGAAAUAAAUUCAAUUGACUUGUUUGCCGCAGCAUUUACAGUUGGCAACAAUUGCAGCAAUGCCAUAAAUCGUUGGACAAGCAUU----------------------- (.(((((((.......))))))))........((((((.(.(((((((((.(((((....)))))(((....)))..))))).))))).))))))..----------------------- ( -20.20) >DroSim_CAF1 9640 97 + 1 GCAAUUUAUAAACGAAAUAAAUUCAAUUGACUUGUUUGCCGCAGCAUUUACAGUUGGCAACAAUUGCAGCAAUGCCAUAAAUCGUUGGACAAGCAUU----------------------- (.(((((((.......))))))))........((((((.(.(((((((((.(((((....)))))(((....)))..))))).))))).))))))..----------------------- ( -20.20) >DroEre_CAF1 11157 97 + 1 GCAAUUUAUAAACGAAAUAAAUUCAAUUGACUUGUUUGCCGCAGCAUUUACAGUUGGCAACAAUUGCAGCAAUGCCAUAAAUCGUUGGACAAGCAUU----------------------- (.(((((((.......))))))))........((((((.(.(((((((((.(((((....)))))(((....)))..))))).))))).))))))..----------------------- ( -20.20) >DroYak_CAF1 9846 97 + 1 GCAAUUUAUAAACGAAAUAAAUUCAAUUGACUUGUUUGCCGCAGCAUUUACAGUUGGCAACAAUUGCAGCAAUGCCAUAAAUCGUUGGACAAGCAUU----------------------- (.(((((((.......))))))))........((((((.(.(((((((((.(((((....)))))(((....)))..))))).))))).))))))..----------------------- ( -20.20) >DroAna_CAF1 61406 120 + 1 GCAAUUUAUAAACGAAAUAAAUUCAAUUGACUUGUUUGCCGCAGCAUUUACAGUUGGCAACAAUUGCAGCAAUGCCAUAAAUCGUUGGACAAGAAAUAUAUACAUACAUACAUGUCCCCU (.(((((((.......))))))))......(((((((..((..(((((..((((((....))))))....))))).......))..)))))))........((((......))))..... ( -19.20) >consensus GCAAUUUAUAAACGAAAUAAAUUCAAUUGACUUGUUUGCCGCAGCAUUUACAGUUGGCAACAAUUGCAGCAAUGCCAUAAAUCGUUGGACAAGCAUU_______________________ (.(((((((.......))))))))......(((((((..((..(((((..((((((....))))))....))))).......))..)))))))........................... (-18.52 = -18.52 + 0.00)

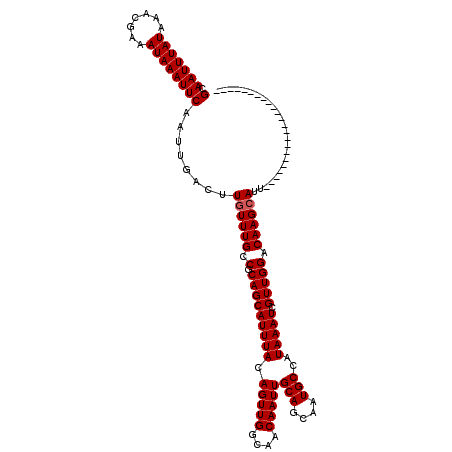

| Location | 4,857,741 – 4,857,838 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.04 |
| Mean single sequence MFE | -21.75 |
| Consensus MFE | -20.00 |
| Energy contribution | -20.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.92 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4857741 97 - 22224390 -----------------------AAUGCUUGUCCAACGAUUUAUGGCAUUGCUGCAAUUGUUGCCAACUGUAAAUGCUGCGGCAAACAAGUCAAUUGAAUUUAUUUCGUUUAUAAAUUGC -----------------------...(((((((((........)))..((((((((....((((.....))))....))))))))))))))......(((((((.......))))))).. ( -20.20) >DroSec_CAF1 9673 97 - 1 -----------------------AAUGCUUGUCCAACGAUUUAUGGCAUUGCUGCAAUUGUUGCCAACUGUAAAUGCUGCGGCAAACAAGUCAAUUGAAUUUAUUUCGUUUAUAAAUUGC -----------------------...(((((((((........)))..((((((((....((((.....))))....))))))))))))))......(((((((.......))))))).. ( -20.20) >DroSim_CAF1 9640 97 - 1 -----------------------AAUGCUUGUCCAACGAUUUAUGGCAUUGCUGCAAUUGUUGCCAACUGUAAAUGCUGCGGCAAACAAGUCAAUUGAAUUUAUUUCGUUUAUAAAUUGC -----------------------...(((((((((........)))..((((((((....((((.....))))....))))))))))))))......(((((((.......))))))).. ( -20.20) >DroEre_CAF1 11157 97 - 1 -----------------------AAUGCUUGUCCAACGAUUUAUGGCAUUGCUGCAAUUGUUGCCAACUGUAAAUGCUGCGGCAAACAAGUCAAUUGAAUUUAUUUCGUUUAUAAAUUGC -----------------------...(((((((((........)))..((((((((....((((.....))))....))))))))))))))......(((((((.......))))))).. ( -20.20) >DroYak_CAF1 9846 97 - 1 -----------------------AAUGCUUGUCCAACGAUUUAUGGCAUUGCUGCAAUUGUUGCCAACUGUAAAUGCUGCGGCAAACAAGUCAAUUGAAUUUAUUUCGUUUAUAAAUUGC -----------------------...(((((((((........)))..((((((((....((((.....))))....))))))))))))))......(((((((.......))))))).. ( -20.20) >DroAna_CAF1 61406 120 - 1 AGGGGACAUGUAUGUAUGUAUAUAUUUCUUGUCCAACGAUUUAUGGCAUUGCUGCAAUUGUUGCCAACUGUAAAUGCUGCGGCAAACAAGUCAAUUGAAUUUAUUUCGUUUAUAAAUUGC ...(((((.((((((....))))))....)))))..(((((((((((.((((((((....((((.....))))....))))))))...........(((.....)))).)))))))))). ( -29.50) >consensus _______________________AAUGCUUGUCCAACGAUUUAUGGCAUUGCUGCAAUUGUUGCCAACUGUAAAUGCUGCGGCAAACAAGUCAAUUGAAUUUAUUUCGUUUAUAAAUUGC ....................................(((((((((((.((((((((....((((.....))))....))))))))...........(((.....)))).)))))))))). (-20.00 = -20.00 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:54 2006