
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,827,517 – 4,827,624 |
| Length | 107 |
| Max. P | 0.586776 |

| Location | 4,827,517 – 4,827,624 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.25 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -20.76 |
| Energy contribution | -21.88 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4827517 107 - 22224390 AGAGCGCUUCA-----AUU-----CGUCUCGUCUCACGUGUCGUAUACGUAAUGUGGGGAAUUUGGUGACUUUGGCCAGC--UUGUGGCAUCUUGCAUCUCCAUAGAGACGAUCAUUAA .(((((.....-----...-----)).)))(((((((((((...))))))..((((((((....(....)....((((..--...))))........)))))))))))))......... ( -31.70) >DroSec_CAF1 33614 112 - 1 AGAGCGCUUCAAUUCAAUU-----CGUCUUGUCUCACGUGUCGUAUACGUAAUGUGGGGAAUUUGGUGACUUUGGCCAGU--UUGUGGCAUCUCGCAUCUCCAUAGAGACGAUCAUUAA ...................-----....(((((((((((((...))))))..((((((((.....((((.....((((..--...))))...)))).)))))))))))))))....... ( -30.40) >DroSim_CAF1 33452 112 - 1 AGAGCGCUUCAAUUCAAUU-----CGUCUCGUCUCACGUGUCGUAUACGUAAUGUGGGGAAUUUGGUGACUUUGGCCAGC--UUGUGGCAUCUCGCAUCUCCAUAGAGACGAUCAUUAA ..................(-----((((((.....((((((...))))))..((((((((.....((((.....((((..--...))))...)))).)))))))))))))))....... ( -32.60) >DroEre_CAF1 43486 105 - 1 AGAUCGCUUCAAUU----------CGUCGCGUCUCACGUGUCGUAUACGUAAUGUGGGGAAUUUGGUGACUUUGGCCAGC--UUGUGGCAU--CGCAUCUCCAUAGAGACGAUCAUUAA .((((((..(....----------.)..))(((((((((((...))))))..((((((((.....((((.....((((..--...)))).)--))).)))))))))))))))))..... ( -34.90) >DroYak_CAF1 34916 117 - 1 AGAUCGCUUCAAUUCAAUUCAUCUCGUCUCGUCUCACGUGUCGUAUACGUAAUGUGGGGAAUUUGGUGGCUUUGGCCAGCAUUUGUGGCAU--CGCAUCUCCAUAGAGACAAUCAUUAA ..............................(((((((((((...))))))..((((((((...((.((((....)))).))..((((....--)))))))))))))))))......... ( -32.60) >DroPer_CAF1 27242 90 - 1 -----------GUUCGUCU-----CGUCUCGUAUCACCUGGCGAAUACGUAAUGUGUGGAAUUUUAUACCCCU-------------ACCAUUCUGCAUCUCCAUGGAGACAAUCAUUAA -----------(((((((.-----.((........))..)))))))...(((((((..((((..((......)-------------)..))))..))((((....))))....))))). ( -18.80) >consensus AGAGCGCUUCAAUUCAAUU_____CGUCUCGUCUCACGUGUCGUAUACGUAAUGUGGGGAAUUUGGUGACUUUGGCCAGC__UUGUGGCAUCUCGCAUCUCCAUAGAGACGAUCAUUAA ............................((((((((((((.....)))))..((((((((.....(((......((((.......))))....))).)))))))))))))))....... (-20.76 = -21.88 + 1.13)

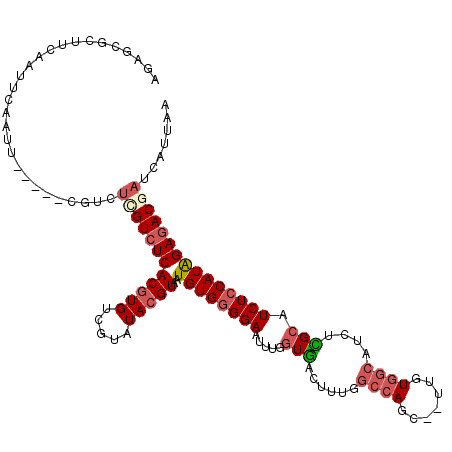

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:45 2006