| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,815,050 – 4,815,210 |
| Length | 160 |
| Max. P | 0.588521 |

| Location | 4,815,050 – 4,815,156 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 96.14 |
| Mean single sequence MFE | -25.86 |
| Consensus MFE | -21.57 |
| Energy contribution | -21.41 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4815050 106 + 22224390 UGCUUUCUUGUCUGUCAUUGAGCCGACAGCACGCGUACCUCCAG-------UAACAACAACCAUACUGACAAUGGCAGCAAUAUCAACAAGUUGGUCAUAACUCGUGACUGAC .......(((.(((((((((.((.....))...........(((-------((..........))))).)))))))))))).........((..(((((.....)))))..)) ( -26.50) >DroSec_CAF1 21593 106 + 1 UGCUUUCUUGUCUGUCAUUGAGCCGACAGCACGCGUACCUCCAG-------UAACAACAACCAUACUGACAAUGGCAGCAAUAUCAACAAGUUGGUCAUAACUCGUGACUGAC .......(((.(((((((((.((.....))...........(((-------((..........))))).)))))))))))).........((..(((((.....)))))..)) ( -26.50) >DroSim_CAF1 20269 106 + 1 UGCUUUCUUGUCUGUCAUUGAGCCGACAGCACGCGUACCUCCAG-------UAACAACAACCAUACUGACAAUGGCAGCAAUAUCAACAAGUUGGUCAUAACUCGUGACUGAC .......(((.(((((((((.((.....))...........(((-------((..........))))).)))))))))))).........((..(((((.....)))))..)) ( -26.50) >DroEre_CAF1 31001 106 + 1 UGCUUUCUUGUCUGUCAUUGAGCCGACAGCACGCGUACCUCCAG-------CAACAACAACCAUACCGACAAUGGCAGCAAUAUCAACAAGUUGGUCAUAACUCGUGACUGAC .......(((.(((((((((.((.....))..((.........)-------).................)))))))))))).........((..(((((.....)))))..)) ( -24.80) >DroYak_CAF1 22091 113 + 1 UGCUUUCUUGUCUGUCAUUGAGCCGACAGCACGCGUACCUCCAACCGCCGACAACAACAACCAUACUGACAAUGGCAGCAAUAUCAACAAGUUGGUCAUAACUCGUGACUGAC .......(((.(((((((((.((.....))..(((..........))).....................)))))))))))).........((..(((((.....)))))..)) ( -25.00) >consensus UGCUUUCUUGUCUGUCAUUGAGCCGACAGCACGCGUACCUCCAG_______UAACAACAACCAUACUGACAAUGGCAGCAAUAUCAACAAGUUGGUCAUAACUCGUGACUGAC .......(((.(((((((((.((.....)).((.(((..........................))))).)))))))))))).........((..(((((.....)))))..)) (-21.57 = -21.41 + -0.16)


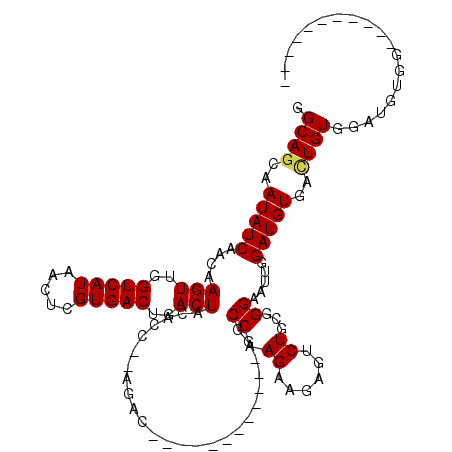
| Location | 4,815,116 – 4,815,210 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 86.44 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -20.58 |
| Energy contribution | -20.66 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4815116 94 + 22224390 GGCAGCAAUAUCAACAAGUUGGUCAUAACUCGUGACUGACUACAAC--AGAC------------GGCCAAGAAGAGUCUGCGGGAAUUGGAUGUGAAUGUGGCUGUGG----------- .(((((.((((..((((((..(((((.....)))))..))).(..(--((((------------...........)))))..)........)))..)))).)))))..----------- ( -30.10) >DroSec_CAF1 21659 94 + 1 GGCAGCAAUAUCAACAAGUUGGUCAUAACUCGUGACUGACUACACC--AGAC------------GGCCAAGAAGAGGCUGCGGGAAUUGGAUGUGACUGUGGAUGUGG----------- .((((..(((((....(((..(((((.....)))))..)))...((--.(.(------------((((.......)))))).)).....)))))..))))........----------- ( -30.50) >DroSim_CAF1 20335 94 + 1 GGCAGCAAUAUCAACAAGUUGGUCAUAACUCGUGACUGACUACACC--AGAC------------GGCCAAGAAGAGGCUGCGGGAAAUGGAUGUGACUGUGGAUGUGG----------- .((((..(((((....(((..(((((.....)))))..)))...((--.(.(------------((((.......)))))).)).....)))))..))))........----------- ( -30.50) >DroEre_CAF1 31067 104 + 1 GGCAGCAAUAUCAACAAGUUGGUCAUAACUCGUGACUGACUACAC---AGAC------------GGCCAAGAAGAGUCUGCGGGAAUUGGAUGUGAUUGUGGAUGUGGAUGGGGAUGUG .(((.(..((((.((((((..(((((.....)))))..))).(((---(..(------------(.((.(((....)))...))...))..))))........))).))))..).))). ( -28.60) >DroYak_CAF1 22164 108 + 1 GGCAGCAAUAUCAACAAGUUGGUCAUAACUCGUGACUGACUACACCACAGACCAGAGAAGACCAGGCCCAGAAGAGUCUGCGGGAAUUGGAUGUGACUGUAGAUGUGA----------- .......(((((.((((((..(((((.....)))))..)))....((((..((((......((((((.(....).))))).)....)))).))))..))).)))))..----------- ( -28.70) >consensus GGCAGCAAUAUCAACAAGUUGGUCAUAACUCGUGACUGACUACACC__AGAC____________GGCCAAGAAGAGUCUGCGGGAAUUGGAUGUGACUGUGGAUGUGG___________ .((((..(((((....(((..(((((.....)))))..))).........................((.((......))...)).....)))))..))))................... (-20.58 = -20.66 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:41 2006