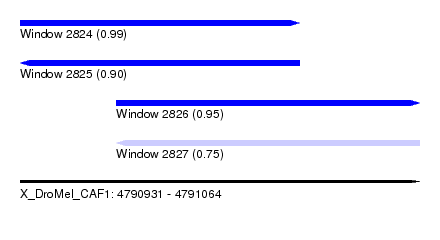
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,790,931 – 4,791,064 |
| Length | 133 |
| Max. P | 0.985985 |

| Location | 4,790,931 – 4,791,024 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 84.53 |
| Mean single sequence MFE | -17.07 |
| Consensus MFE | -15.50 |
| Energy contribution | -15.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.985985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4790931 93 + 22224390 AGAAAUCAUAUUCAAAUUGCAUAUGCACUUCGAGCGUAGGAAAAUCUGUUUUUCCUACCAUUCAGCUCUGCUGCCUUUUCAAAUUUUCAAUCC .((((............(((....)))....(((((((((((((.....)))))))))......))))........))))............. ( -18.20) >DroSec_CAF1 36721 75 + 1 AGAAAUCAUAUUCAAAUUGCAUAUGCACUUCGAGCGUAGGAAAAUCUGUUUUUCCUACCAU-----------------UA-AAUUUUCAAUCC .((((.....(((.((.(((....))).)).))).(((((((((.....)))))))))...-----------------..-...))))..... ( -15.50) >DroSim_CAF1 23174 92 + 1 AGAAAUCAUAUUCAAAUUGCAUAUGCACUUCGAGCGUAGGAAAAUCUGUUUUUCCUACCAUUCAGCACUGCUGCCCAUUC-AAUUUUCAGUCC .((((.....(((.((.(((....))).)).))).(((((((((.....)))))))))....((((...)))).......-...))))..... ( -17.50) >consensus AGAAAUCAUAUUCAAAUUGCAUAUGCACUUCGAGCGUAGGAAAAUCUGUUUUUCCUACCAUUCAGC_CUGCUGCC__UUC_AAUUUUCAAUCC .((((.....(((.((.(((....))).)).))).(((((((((.....)))))))))..........................))))..... (-15.50 = -15.50 + 0.00)



| Location | 4,790,931 – 4,791,024 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 84.53 |
| Mean single sequence MFE | -20.70 |
| Consensus MFE | -18.20 |
| Energy contribution | -18.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.900828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4790931 93 - 22224390 GGAUUGAAAAUUUGAAAAGGCAGCAGAGCUGAAUGGUAGGAAAAACAGAUUUUCCUACGCUCGAAGUGCAUAUGCAAUUUGAAUAUGAUUUCU .............((((...((((...))))....(((((((((.....)))))))))..(((((.(((....))).)))))......)))). ( -21.00) >DroSec_CAF1 36721 75 - 1 GGAUUGAAAAUU-UA-----------------AUGGUAGGAAAAACAGAUUUUCCUACGCUCGAAGUGCAUAUGCAAUUUGAAUAUGAUUUCU .....((((...-..-----------------...(((((((((.....)))))))))..(((((.(((....))).)))))......)))). ( -18.20) >DroSim_CAF1 23174 92 - 1 GGACUGAAAAUU-GAAUGGGCAGCAGUGCUGAAUGGUAGGAAAAACAGAUUUUCCUACGCUCGAAGUGCAUAUGCAAUUUGAAUAUGAUUUCU ............-....(((((((...))).....(((((((((.....)))))))))))))(((((.(((((.(.....).)))))))))). ( -22.90) >consensus GGAUUGAAAAUU_GAA__GGCAGCAG_GCUGAAUGGUAGGAAAAACAGAUUUUCCUACGCUCGAAGUGCAUAUGCAAUUUGAAUAUGAUUUCU .....((((..........................(((((((((.....)))))))))..(((((.(((....))).)))))......)))). (-18.20 = -18.20 + -0.00)


| Location | 4,790,963 – 4,791,064 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 83.11 |
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -19.29 |
| Energy contribution | -19.73 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.949971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4790963 101 + 22224390 AGCGUAGGAAAAUCUGUUUUUCCUACCAUUCAGCUCUGCUGCCUUUUCAAAUUUUCAAUCCAUUUUCCGGACUACGAAUGUUUUCCAGCAUGAUCGGAAUC .(.(((((((((.....))))))))))((((.(.(((((((.................(((.......)))....(((....)))))))).)).).)))). ( -21.70) >DroSec_CAF1 36753 83 + 1 AGCGUAGGAAAAUCUGUUUUUCCUACCAU-----------------UA-AAUUUUCAAUCCAUUUUCCGGAUUCCGGAUGUUUUGCAGCAUGAUCGGAAUC .(.(((((((((.....))))))))))..-----------------..-....................((((((((((((......))))..)))))))) ( -22.00) >DroSim_CAF1 23206 100 + 1 AGCGUAGGAAAAUCUGUUUUUCCUACCAUUCAGCACUGCUGCCCAUUC-AAUUUUCAGUCCAUUUUCCGGAUUCCGGAUGUUUUCCAGCAUGAUCGGAAUC .(.(((((((((.....))))))))))...((((...)))).......-....................((((((((((((......))))..)))))))) ( -24.40) >consensus AGCGUAGGAAAAUCUGUUUUUCCUACCAUUCAGC_CUGCUGCC__UUC_AAUUUUCAAUCCAUUUUCCGGAUUCCGGAUGUUUUCCAGCAUGAUCGGAAUC .(.(((((((((.....))))))))))..........................................((((((((((((......))))..)))))))) (-19.29 = -19.73 + 0.45)


| Location | 4,790,963 – 4,791,064 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 83.11 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -17.25 |
| Energy contribution | -17.37 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4790963 101 - 22224390 GAUUCCGAUCAUGCUGGAAAACAUUCGUAGUCCGGAAAAUGGAUUGAAAAUUUGAAAAGGCAGCAGAGCUGAAUGGUAGGAAAAACAGAUUUUCCUACGCU ..(((((.......)))))..(((((.(((((((.....)))))))............(((......))))))))(((((((((.....)))))))))... ( -27.00) >DroSec_CAF1 36753 83 - 1 GAUUCCGAUCAUGCUGCAAAACAUCCGGAAUCCGGAAAAUGGAUUGAAAAUU-UA-----------------AUGGUAGGAAAAACAGAUUUUCCUACGCU (((....)))..((..(((..(((((((...)))))...))..)))......-..-----------------...(((((((((.....))))))))))). ( -19.80) >DroSim_CAF1 23206 100 - 1 GAUUCCGAUCAUGCUGGAAAACAUCCGGAAUCCGGAAAAUGGACUGAAAAUU-GAAUGGGCAGCAGUGCUGAAUGGUAGGAAAAACAGAUUUUCCUACGCU ...((((.(((..(((((.....)))))..((((.....))))........)-)).))))((((...))))....(((((((((.....)))))))))... ( -26.40) >consensus GAUUCCGAUCAUGCUGGAAAACAUCCGGAAUCCGGAAAAUGGAUUGAAAAUU_GAA__GGCAGCAG_GCUGAAUGGUAGGAAAAACAGAUUUUCCUACGCU .....(((((....((.....))((((.....)))).....))))).............................(((((((((.....)))))))))... (-17.25 = -17.37 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:29 2006