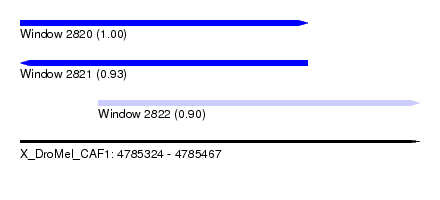
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,785,324 – 4,785,467 |
| Length | 143 |
| Max. P | 0.995843 |

| Location | 4,785,324 – 4,785,427 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 82.18 |
| Mean single sequence MFE | -34.89 |
| Consensus MFE | -21.76 |
| Energy contribution | -25.64 |
| Covariance contribution | 3.88 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4785324 103 + 22224390 AGUGUGAUUAU--GUAUGCAUGUGUGUCUGCAUCAAAUCACACACUUGCAGCGCUCAUGCACACGCACUUGUACUUGUGCGUGGCUCACAUUUCCAUAGCCAGCC (((((((....--...((((((.(((.(((((..............)))))))).))))))(((((((........)))))))..)))))))............. ( -37.44) >DroSec_CAF1 31227 105 + 1 AGUGUGAUUAUAUGUGUGUGUGUGUGUCUGCAUCAAAUCACACACUUGCAGCGCUCAUGCACACGCACUUGUACUUGUGCGUGGCUCACAUUUCCAUAGCCAGCC (((((((((((..(((((((((.(((.(((((..............)))))))).)))))))))((((........)))))))).)))))))............. ( -36.64) >DroEre_CAF1 13784 80 + 1 AGUGUG--UAU--GUGUG----UGUGUGUGCAUCAAAUC-----------------AUGCACACGCACUUGUACUUGUGCGUGGCUCACAUUUCCAUAGCCCCCC ..((((--...--(((((----.((((((((((......-----------------))))))))((((........))))...)).)))))...))))....... ( -27.30) >DroYak_CAF1 14960 102 + 1 AGUGUAUGCAU--GUGUGUA-GUGUGUGUGCAUCAAAUCACACACUUGCAGCGCUCAUGCACACGCACUUGUACUUGUGCGUGUCUCACAUUUCCAUAGCCCCCA .(((...((((--(.((((.-(((((((((........)))))))..)).)))).)))))((((((((........))))))))..)))................ ( -38.20) >consensus AGUGUGAUUAU__GUGUGUA_GUGUGUCUGCAUCAAAUCACACACUUGCAGCGCUCAUGCACACGCACUUGUACUUGUGCGUGGCUCACAUUUCCAUAGCCACCC (((((((((((..(((((((((.(((.(((((..............)))))))).)))))))))((((........)))))))).)))))))............. (-21.76 = -25.64 + 3.88)


| Location | 4,785,324 – 4,785,427 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 82.18 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -19.58 |
| Energy contribution | -21.20 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4785324 103 - 22224390 GGCUGGCUAUGGAAAUGUGAGCCACGCACAAGUACAAGUGCGUGUGCAUGAGCGCUGCAAGUGUGUGAUUUGAUGCAGACACACAUGCAUAC--AUAAUCACACU ((((.((.((....)))).))))((((((........))))))((((....)))).......(((((((((((((((........))))).)--).)))))))). ( -39.40) >DroSec_CAF1 31227 105 - 1 GGCUGGCUAUGGAAAUGUGAGCCACGCACAAGUACAAGUGCGUGUGCAUGAGCGCUGCAAGUGUGUGAUUUGAUGCAGACACACACACACACAUAUAAUCACACU ((((.((.((....)))).))))((((((........))))))((((....)))).....(((((((.((((...)))))))))))................... ( -34.70) >DroEre_CAF1 13784 80 - 1 GGGGGGCUAUGGAAAUGUGAGCCACGCACAAGUACAAGUGCGUGUGCAU-----------------GAUUUGAUGCACACACA----CACAC--AUA--CACACU ....(..((((....((((......((((........))))((((((((-----------------......)))))))).))----))..)--)))--..)... ( -24.30) >DroYak_CAF1 14960 102 - 1 UGGGGGCUAUGGAAAUGUGAGACACGCACAAGUACAAGUGCGUGUGCAUGAGCGCUGCAAGUGUGUGAUUUGAUGCACACACAC-UACACAC--AUGCAUACACU ................(((..((((((((........))))))))(((((.((...))..(((((((........)))))))..-......)--))))...))). ( -33.10) >consensus GGCGGGCUAUGGAAAUGUGAGCCACGCACAAGUACAAGUGCGUGUGCAUGAGCGCUGCAAGUGUGUGAUUUGAUGCACACACAC_UACACAC__AUAAUCACACU ...............((((.(((((((((........))))))).)).............(((((((.((((...))))))))))).)))).............. (-19.58 = -21.20 + 1.62)


| Location | 4,785,352 – 4,785,467 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.61 |
| Mean single sequence MFE | -22.34 |
| Consensus MFE | -16.15 |
| Energy contribution | -16.15 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4785352 115 + 22224390 CAUCAAAUCACACACUUGCAGCGCUCAUGCACACGCACUUGUACUUGUGCGUGGCUCACAUUUCCAUAGCCAGCCGCUUCCCCCUCCACGAUCAGUGUUGCUCUUCACCUGCAGC ...............((((((.......((((((((((........))))((((((..(.........)..)))))).................))).))).......)))))). ( -27.24) >DroSec_CAF1 31257 115 + 1 CAUCAAAUCACACACUUGCAGCGCUCAUGCACACGCACUUGUACUUGUGCGUGGCUCACAUUUCCAUAGCCAGCCGCUUCCCCCUCCACGAUCAUUGUUGCUCAUCACCUGCAGC ...............((((((.......(((.((((((........))))((((((..(.........)..))))))...................))))).......)))))). ( -23.64) >DroEre_CAF1 13806 93 + 1 CAUCAAAUC-----------------AUGCACACGCACUUGUACUUGUGCGUGGCUCACAUUUCCAUAGCCCCCCGCU-CCCUCUCC----UUGUUGUUUCUCUUCACCUGCAGC .........-----------------..((.(((((((........)))))))))............(((.....)))-........----..(((((............))))) ( -17.30) >DroYak_CAF1 14987 107 + 1 CAUCAAAUCACACACUUGCAGCGCUCAUGCACACGCACUUGUACUUGUGCGUGUCUCACAUUUCCAUAGCCCCCAGCC-CCCUCUCU----UUGUU---ACUUUUCACCUGCAGC ...............((((((.((....))((((((((........))))))))........................-........----.....---.........)))))). ( -21.20) >consensus CAUCAAAUCACACACUUGCAGCGCUCAUGCACACGCACUUGUACUUGUGCGUGGCUCACAUUUCCAUAGCCACCCGCU_CCCCCUCC____UCAUUGUUGCUCUUCACCUGCAGC ...........................(((((((((((........)))))))...............((.....))................................)))).. (-16.15 = -16.15 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:25 2006