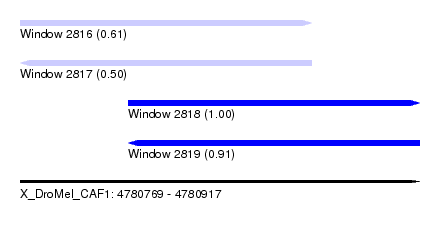
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,780,769 – 4,780,917 |
| Length | 148 |
| Max. P | 0.999550 |

| Location | 4,780,769 – 4,780,877 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.43 |
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -22.51 |
| Energy contribution | -23.07 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4780769 108 + 22224390 GCCAACUGACGAAACUGACGAAAAUCGACUAGAGGUGGUGGAGA---------AACAUCGAUGCCAAACAUCGAUAUUUUCGAAAAAUCACAUAAAUUGGUG---UUUUUUGAUGAAUAU ..(((..(((........((.....))(((((..(((((...((---------((.(((((((.....)))))))..)))).....))))).....))))))---))..)))........ ( -20.00) >DroSec_CAF1 20359 117 + 1 GCCAACUGGCGAAACUGACGAAAAUCGACUAGAGGUGGAGAAAAAUUGAUGCCAACAUCGAUGUCAAACAUCGAUAUUUUCGAAGAAUCACAAACAUCGGUG---UUUUUCGAGGAAUCA (((....))).....((((.....((.((.....)).)).....(((((((....)))))))))))......(((.(((((((((((.(((........)))---)))))))))))))). ( -31.00) >DroSim_CAF1 14495 120 + 1 GCCAACUGGCGAAACUGACGAAAAUCGACUAGAGGUGGAGAAAAAUUGAUGCCAACAUCGAUGUCAAACAUCGAUAUUUUCGAAGAAUCACAAACAUCGGUUUGUUUUUUCGAGGAAUCA (((....))).....((((.....((.((.....)).)).....(((((((....)))))))))))......(((.(((((((((((..((((((....)))))))))))))))))))). ( -32.80) >consensus GCCAACUGGCGAAACUGACGAAAAUCGACUAGAGGUGGAGAAAAAUUGAUGCCAACAUCGAUGUCAAACAUCGAUAUUUUCGAAGAAUCACAAACAUCGGUG___UUUUUCGAGGAAUCA .(((.(((((((............))).))))...)))..................(((((((.....))))))).(((((((((((.(((........)))...))))))))))).... (-22.51 = -23.07 + 0.56)

| Location | 4,780,769 – 4,780,877 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.43 |
| Mean single sequence MFE | -26.27 |
| Consensus MFE | -21.26 |
| Energy contribution | -20.60 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4780769 108 - 22224390 AUAUUCAUCAAAAAA---CACCAAUUUAUGUGAUUUUUCGAAAAUAUCGAUGUUUGGCAUCGAUGUU---------UCUCCACCACCUCUAGUCGAUUUUCGUCAGUUUCGUCAGUUGGC ...............---..((((((.(((.((((...((((((((((((((.....))))))))).---------..((.((........)).)).)))))..)))).))).)))))). ( -22.20) >DroSec_CAF1 20359 117 - 1 UGAUUCCUCGAAAAA---CACCGAUGUUUGUGAUUCUUCGAAAAUAUCGAUGUUUGACAUCGAUGUUGGCAUCAAUUUUUCUCCACCUCUAGUCGAUUUUCGUCAGUUUCGCCAGUUGGC ((((.(((((((.((---(((........))).)).))))).((((((((((.....)))))))))))).)))).................(((((((..((.......))..))))))) ( -26.90) >DroSim_CAF1 14495 120 - 1 UGAUUCCUCGAAAAAACAAACCGAUGUUUGUGAUUCUUCGAAAAUAUCGAUGUUUGACAUCGAUGUUGGCAUCAAUUUUUCUCCACCUCUAGUCGAUUUUCGUCAGUUUCGCCAGUUGGC ((((.(((((((...((((((....)))))).....))))).((((((((((.....)))))))))))).)))).................(((((((..((.......))..))))))) ( -29.70) >consensus UGAUUCCUCGAAAAA___CACCGAUGUUUGUGAUUCUUCGAAAAUAUCGAUGUUUGACAUCGAUGUUGGCAUCAAUUUUUCUCCACCUCUAGUCGAUUUUCGUCAGUUUCGCCAGUUGGC ....................(((((....((((..((.((((((((((((((.....))))))))))....................((.....))..))))..))..))))..))))). (-21.26 = -20.60 + -0.66)


| Location | 4,780,809 – 4,780,917 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.40 |
| Mean single sequence MFE | -26.13 |
| Consensus MFE | -17.24 |
| Energy contribution | -17.47 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.66 |
| SVM decision value | 3.71 |
| SVM RNA-class probability | 0.999550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4780809 108 + 22224390 GAGA---------AACAUCGAUGCCAAACAUCGAUAUUUUCGAAAAAUCACAUAAAUUGGUG---UUUUUUGAUGAAUAUUCAUUUUUGAUGUAUAUUCAAAUUUCUGAUGAAUCAUUGU .(((---------((.(((((((.....)))))))....((((((((.(((........)))---))))))))(((((((.((((...)))).)))))))..)))))............. ( -23.90) >DroSec_CAF1 20399 116 + 1 AAAAAUUGAUGCCAACAUCGAUGUCAAACAUCGAUAUUUUCGAAGAAUCACAAACAUCGGUG---UUUUUCGAGGAAUCAUCGUAUAAAUUGUAAGUACAAAUUAAUUUUGCAU-AUCUU .(((((((((....(((((((((.....))))))).(((((((((((.(((........)))---)))))))))))...................))....)))))))))....-..... ( -26.20) >DroSim_CAF1 14535 119 + 1 AAAAAUUGAUGCCAACAUCGAUGUCAAACAUCGAUAUUUUCGAAGAAUCACAAACAUCGGUUUGUUUUUUCGAGGAAUCAUCGUAUAAAUUGUAAGAACAAAUUAAUUUUGCAU-AUCUU .......((((.....(((((((.....))))))).(((((((((((..((((((....)))))))))))))))))..))))........(((((((.........))))))).-..... ( -28.30) >consensus AAAAAUUGAUGCCAACAUCGAUGUCAAACAUCGAUAUUUUCGAAGAAUCACAAACAUCGGUG___UUUUUCGAGGAAUCAUCGUAUAAAUUGUAAGUACAAAUUAAUUUUGCAU_AUCUU ................(((((((.....))))))).(((((((((((.(((........)))...)))))))))))............................................ (-17.24 = -17.47 + 0.23)

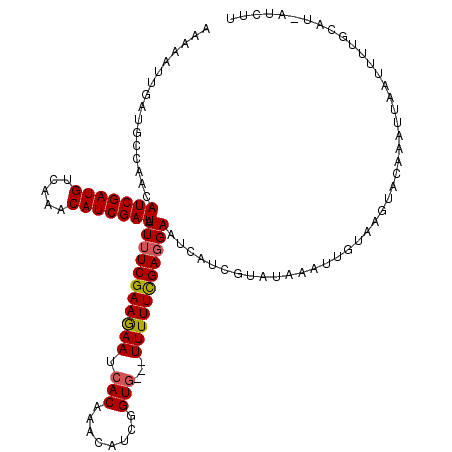
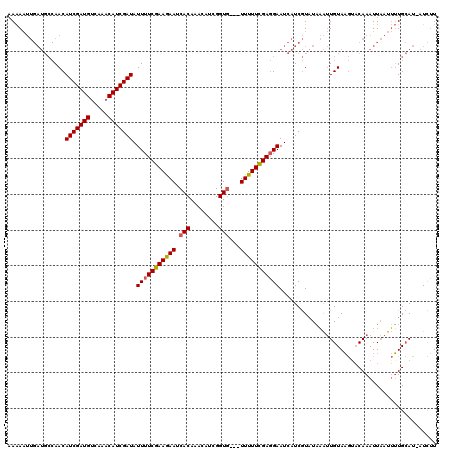
| Location | 4,780,809 – 4,780,917 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.40 |
| Mean single sequence MFE | -24.53 |
| Consensus MFE | -13.39 |
| Energy contribution | -14.17 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4780809 108 - 22224390 ACAAUGAUUCAUCAGAAAUUUGAAUAUACAUCAAAAAUGAAUAUUCAUCAAAAAA---CACCAAUUUAUGUGAUUUUUCGAAAAUAUCGAUGUUUGGCAUCGAUGUU---------UCUC ....................(((((((.(((.....))).)))))))((.(((((---(((........))).))))).))(((((((((((.....))))))))))---------)... ( -21.50) >DroSec_CAF1 20399 116 - 1 AAGAU-AUGCAAAAUUAAUUUGUACUUACAAUUUAUACGAUGAUUCCUCGAAAAA---CACCGAUGUUUGUGAUUCUUCGAAAAUAUCGAUGUUUGACAUCGAUGUUGGCAUCAAUUUUU .....-.((((((.....))))))................((((.(((((((.((---(((........))).)).))))).((((((((((.....)))))))))))).))))...... ( -24.60) >DroSim_CAF1 14535 119 - 1 AAGAU-AUGCAAAAUUAAUUUGUUCUUACAAUUUAUACGAUGAUUCCUCGAAAAAACAAACCGAUGUUUGUGAUUCUUCGAAAAUAUCGAUGUUUGACAUCGAUGUUGGCAUCAAUUUUU ((((.-..(((((.....))))))))).............((((.(((((((...((((((....)))))).....))))).((((((((((.....)))))))))))).))))...... ( -27.50) >consensus AAGAU_AUGCAAAAUUAAUUUGUACUUACAAUUUAUACGAUGAUUCCUCGAAAAA___CACCGAUGUUUGUGAUUCUUCGAAAAUAUCGAUGUUUGACAUCGAUGUUGGCAUCAAUUUUU .......((((((.....)))))).............(((.((.((..((((..............)))).)).)).)))..((((((((((.....))))))))))............. (-13.39 = -14.17 + 0.78)

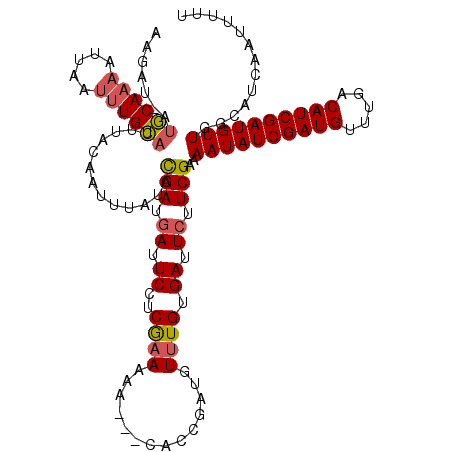
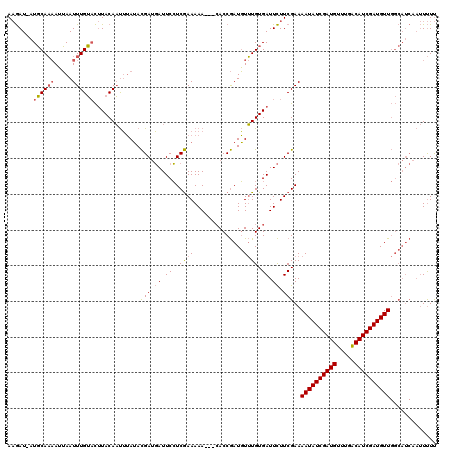
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:23 2006