
| Sequence ID | X_DroMel_CAF1 |
|---|---|
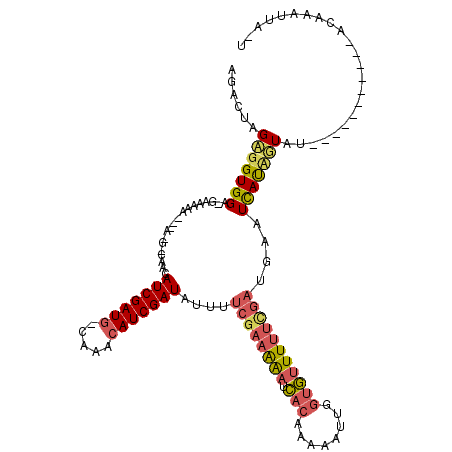
| Location | 4,777,341 – 4,777,440 |
| Length | 99 |
| Max. P | 0.923799 |

| Location | 4,777,341 – 4,777,440 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 58.91 |
| Mean single sequence MFE | -22.96 |
| Consensus MFE | -10.40 |
| Energy contribution | -12.30 |
| Covariance contribution | 1.90 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.45 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4777341 99 + 22224390 GGACUAGAGGUGGU-GGAGA---------AACAUCGAUGCCAAACAUCGAUAUUUUCGAAAAAUCACAUAAAUUGGUGUUUUUUGAUGAAUCAUUUUAU-----------UCAAAUUUUU ......((((((((-.....---------...(((((((.....)))))))....((((((((.(((........)))))))))))...))))))))..-----------.......... ( -20.20) >DroSec_CAF1 14165 119 + 1 CGACUAGAGGUGGA-GAAAAAUUGAUGCCAACAUCGAUGUCAAACAUCGAUAUUUUCGAAGAAUCACAAACAUCGGUGUUUUUCGAGGAAUCAUCGUAUAAAUUGUAAGUACAAAUUAAU (((.....((((..-((...(((((((....))))))).))...))))(((.(((((((((((.(((........))))))))))))))))).)))..(((.((((....)))).))).. ( -27.80) >DroWil_CAF1 12799 103 + 1 AAAGUAGCGGUGGAAGAAACAACAAGGGCACUAUCGAU-----ACAUCGAUAAUUUUGCAAGUUUUCGAUAAAUACUAUUUGUGCCUGAAUUACAGUUU-----------CCAAAUUA-U ..........((((((.........((((((((((((.-----...))))))........(((...........)))....)))))).........)))-----------))).....-. ( -20.87) >consensus AGACUAGAGGUGGA_GAAAAA___A_G_CAACAUCGAUG_CAAACAUCGAUAUUUUCGAAAAAUCACAAAAAUUGGUGUUUUUCGAUGAAUCAUAGUAU___________ACAAAUUA_U ......(((((((...................(((((((.....)))))))....((((((((.(((........)))))))))))....)))))))....................... (-10.40 = -12.30 + 1.90)


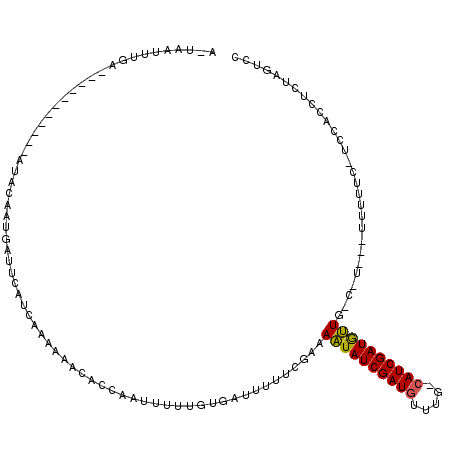
| Location | 4,777,341 – 4,777,440 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 58.91 |
| Mean single sequence MFE | -20.17 |
| Consensus MFE | -10.46 |
| Energy contribution | -10.13 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4777341 99 - 22224390 AAAAAUUUGA-----------AUAAAAUGAUUCAUCAAAAAACACCAAUUUAUGUGAUUUUUCGAAAAUAUCGAUGUUUGGCAUCGAUGUU---------UCUCC-ACCACCUCUAGUCC .......(((-----------((......)))))((.((((((((........))).))))).))(((((((((((.....))))))))))---------)....-.............. ( -17.60) >DroSec_CAF1 14165 119 - 1 AUUAAUUUGUACUUACAAUUUAUACGAUGAUUCCUCGAAAAACACCGAUGUUUGUGAUUCUUCGAAAAUAUCGAUGUUUGACAUCGAUGUUGGCAUCAAUUUUUC-UCCACCUCUAGUCG ..(((.((((....)))).))).....((((.(((((((.(((((........))).)).))))).((((((((((.....)))))))))))).)))).......-.............. ( -23.60) >DroWil_CAF1 12799 103 - 1 A-UAAUUUGG-----------AAACUGUAAUUCAGGCACAAAUAGUAUUUAUCGAAAACUUGCAAAAUUAUCGAUGU-----AUCGAUAGUGCCCUUGUUGUUUCUUCCACCGCUACUUU .-.....(((-----------((....((((...(((((.....(((.............))).....((((((...-----.)))))))))))...))))....))))).......... ( -19.32) >consensus A_UAAUUUGA___________AUACAAUGAUUCAUCAAAAAACACCAAUUUUUGUGAUUUUUCGAAAAUAUCGAUGUUUG_CAUCGAUGUUG_C_U___UUUUUC_UCCACCUCUAGUCC ..................................................................((((((((((.....))))))))))............................. (-10.46 = -10.13 + -0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:14 2006