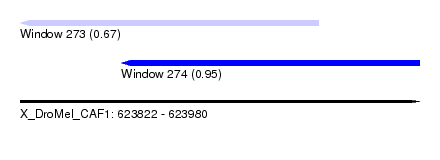
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 623,822 – 623,980 |
| Length | 158 |
| Max. P | 0.949208 |

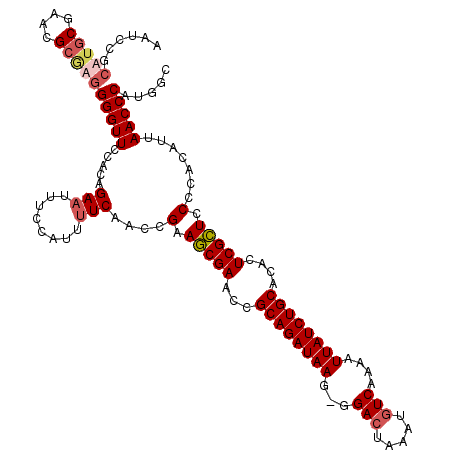
| Location | 623,822 – 623,940 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.73 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -20.88 |
| Energy contribution | -22.00 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 623822 118 - 22224390 UUUCAACCGAAGCGAACCGCAGAUAAG-GGA-UAAAUGUCAAAAUUAUCUGCACACUCGCUCCCCACAUUAACCCAUGCCGUACGCACUAUCGUGCAACUGACCAAACCCAAGGUGAAGG (((((.(((.(((((...((((((((.-.((-(....)))....))))))))....))))).).....................((((....))))................))))))). ( -28.20) >DroSec_CAF1 8318 119 - 1 UUUCAACCGAAGCGAACCGCAGAUAAG-GGACUAAAUGUCAAAAUUAUCUGCACACUCGCUCCUCACAUUAACCCAUGGCGUACGCACUGUCGCGCAACUGACCAAACACAAGGUGGAGG ..........(((((...((((((((.-.(((.....)))....))))))))....)))))((((.............((((((.....)).)))).....(((........))).)))) ( -31.50) >DroSim_CAF1 2578 120 - 1 UUUCAACCGAAGCGAACCGCAGAUAAGGGGACUAAAUGUCAAAAUUAUCUGCACACUCGCUCCUCACAUUAACCCAUGGCGUACGCACUGUCGUGCAACUGACCAAACCCAAGGUGGAGG ..........(((((...((((((((...(((.....)))....))))))))....)))))((((.((((......(((((...((((....))))...)).))).......)))))))) ( -33.82) >DroEre_CAF1 8636 110 - 1 U-UCAGCCGAAACGAACUGCAGAUAAG-GGACUAAAUGUCAAAAUUAUCUGCACACUCGUUCCCCAUAUUAACCCAUGGCGUGCGCAUUGUCCCUCAGCUGCCCAAGCCCAA-------- .-......(.(((((..(((((((((.-.(((.....)))....)))))))))...))))).)..............(((.((.((((((.....))).))).)).)))...-------- ( -28.80) >consensus UUUCAACCGAAGCGAACCGCAGAUAAG_GGACUAAAUGUCAAAAUUAUCUGCACACUCGCUCCCCACAUUAACCCAUGGCGUACGCACUGUCGCGCAACUGACCAAACCCAAGGUGGAGG .....((((.(((((...((((((((...(((.....)))....))))))))....))))).).................(((((......)))))................)))..... (-20.88 = -22.00 + 1.12)

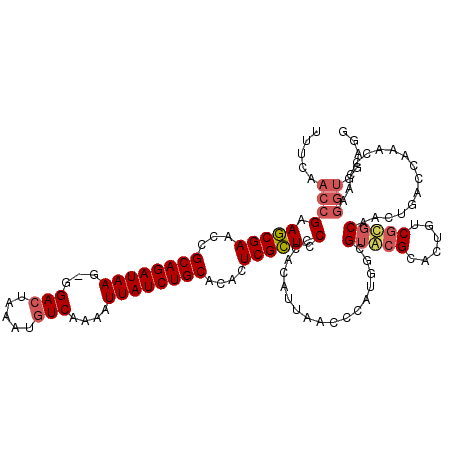
| Location | 623,862 – 623,980 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.61 |
| Mean single sequence MFE | -32.60 |
| Consensus MFE | -27.02 |
| Energy contribution | -28.27 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 623862 118 - 22224390 AAUCCGCAUGCGAACGCAAGGGGUUCCUCAGAAUUUCCAUUUUCAACCGAAGCGAACCGCAGAUAAG-GGA-UAAAUGUCAAAAUUAUCUGCACACUCGCUCCCCACAUUAACCCAUGCC .....((((((....))..((((.......(((........)))......(((((...((((((((.-.((-(....)))....))))))))....)))))))))..........)))). ( -31.00) >DroSec_CAF1 8358 119 - 1 AAUCCGCUUGCGAACGCGAGGGGUUCCACAGAAUUUCCAUUUUCAACCGAAGCGAACCGCAGAUAAG-GGACUAAAUGUCAAAAUUAUCUGCACACUCGCUCCUCACAUUAACCCAUGGC ...((((((((....)))))(((((.....(((........)))....(((((((...((((((((.-.(((.....)))....))))))))....)))))..)).....))))).))). ( -34.60) >DroSim_CAF1 2618 120 - 1 AAUCCGCUUGCGAACGCGAGGGGUUCCACAGAAUUUCCAUUUUCAACCGAAGCGAACCGCAGAUAAGGGGACUAAAUGUCAAAAUUAUCUGCACACUCGCUCCUCACAUUAACCCAUGGC ...((((((((....)))))(((((.....(((........)))....(((((((...((((((((...(((.....)))....))))))))....)))))..)).....))))).))). ( -34.90) >DroEre_CAF1 8668 116 - 1 AAUCCGCAGGCGAACG--AAGGGUUCCUCAGAAUUUCCAUU-UCAGCCGAAACGAACUGCAGAUAAG-GGACUAAAUGUCAAAAUUAUCUGCACACUCGUUCCCCAUAUUAACCCAUGGC ........(((((((.--....))))....(((.......)-)).)))(.(((((..(((((((((.-.(((.....)))....)))))))))...))))).)((((........)))). ( -29.90) >consensus AAUCCGCAUGCGAACGCGAGGGGUUCCACAGAAUUUCCAUUUUCAACCGAAGCGAACCGCAGAUAAG_GGACUAAAUGUCAAAAUUAUCUGCACACUCGCUCCCCACAUUAACCCAUGGC ......(((((....)))))(((((.....(((........)))....(.(((((...((((((((...(((.....)))....))))))))....))))).).......)))))..... (-27.02 = -28.27 + 1.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:02 2006