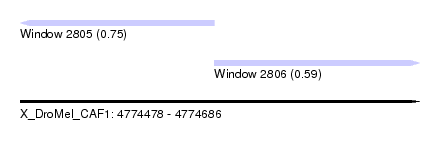
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,774,478 – 4,774,686 |
| Length | 208 |
| Max. P | 0.747493 |

| Location | 4,774,478 – 4,774,579 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -31.35 |
| Consensus MFE | -25.17 |
| Energy contribution | -25.83 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
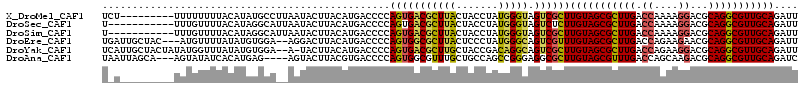
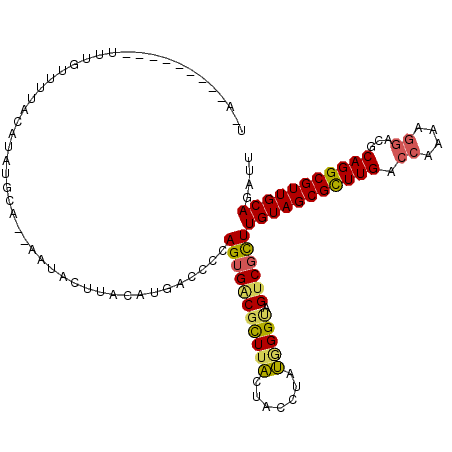
>X_DroMel_CAF1 4774478 101 - 22224390 AAAAUAGUAGCU-------UAUCGUUUUGCGCACAUUACAGGUGGUGGAUGCCCGUGCUGGACCCGGCACCAAGUCCAUGGCCAAACAGAAGAAGCGCGACAACGCAG ............-------...((((((((((........(((.((((((....((((((....))))))...)))))).)))...(....)..)))))).))))... ( -34.70) >DroPse_CAF1 8662 97 - 1 ACUUUACUAACUCUCUGUUU-----------CCUGUAAAAGGUGGUGGAUGCCCGAGCUGGUCCCGGCACCAAGUCCCUGGCCAAACAGAAAAAGCGCGAGAACGCAG ...........(((((((((-----------.((((....(((.(.((((....(.((((....)))).)...)))).).)))..))))...))))).))))...... ( -24.00) >DroSim_CAF1 9844 108 - 1 AAAAUACUAACCCGCUUUCUCUCGCUUUGCGCACAUUGCAGGUGGUGGAUGCCCGUGCUGGACCCGGCACCAAGUCCAUGGCCAAACAGAAGAAGCGCGACAACGCAG .............((((..(((.((..((....))..)).(((.((((((....((((((....))))))...)))))).)))....)))..))))(((....))).. ( -39.50) >DroEre_CAF1 7929 108 - 1 GCAAAACUAAUCCUCUGUUUUACCCUUUGCGCACAUAACAGGUGGUGGAUGCCCGUGCUGGACCCGGCACCAAGUCCAUGGCCAAACAAAAGAAGCGCGACAACGCAG (((((((.........))))).....((((((........(((.((((((....((((((....))))))...)))))).)))...(....)..))))))....)).. ( -35.30) >DroYak_CAF1 8679 97 - 1 GUAAAACUAAUCCUCUCUCU-----------CACAUGACAGGUGGUGGAUGCUCGUGCUGGACCCGGCACCAAGUCCAUGGCCAAACAAAAGAAGCGUGACAACGCAG ....................-----------.........(((.((((((....((((((....))))))...)))))).)))...........((((....)))).. ( -30.60) >DroPer_CAF1 9158 97 - 1 ACUUUACUAACUCUCUGUUU-----------CCUGUAAAAGGUGGUGGAUGCCCGAGCUGGUCCCGGCACCAAGUCCCUGGCCAAACAGAAAAAGCGCGAGAACGCAG ...........(((((((((-----------.((((....(((.(.((((....(.((((....)))).)...)))).).)))..))))...))))).))))...... ( -24.00) >consensus ACAAUACUAACCCUCUGUUU___________CACAUAACAGGUGGUGGAUGCCCGUGCUGGACCCGGCACCAAGUCCAUGGCCAAACAGAAGAAGCGCGACAACGCAG ........................................(((.((((((....((((((....))))))...)))))).)))...........(((......))).. (-25.17 = -25.83 + 0.67)


| Location | 4,774,579 – 4,774,686 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.03 |
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -21.94 |
| Energy contribution | -21.70 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4774579 107 + 22224390 UCU---------UUUUUUUUACAUAUGCCUUAAUACUUACAUGACCCCAGUGACGCUUACUACCUAUGGGUAGUCGCUUGUAGCGCUUGACCAAAAGGACGCAGGCGUUGCAGAUU ...---------........................((((.((....)))))).((..((((((....)))))).))((((((((((((.((....))...))))))))))))... ( -28.00) >DroSec_CAF1 11172 105 + 1 U-----------UUUGUUUUACAUAGGCAUUAAUACUUACAUGACCCCAGUGACGCUUACUACCUAUGGGUAGUCUCUUGUAGCGCUUGACCAAAAGGACGCAGGCGUUGCAGAUU .-----------...((.((((...(((((..........))).))...)))).))..((((((....))))))...((((((((((((.((....))...))))))))))))... ( -29.20) >DroSim_CAF1 9952 105 + 1 U-----------UUUGUUUUACAUAGGCAUUAAUACUUACAUGACCCCAGUGACGCUUACUACCUAUGGGUAGUCGCUUGUAGCGCUUGACCAAAAGGACGCAGGCGUUGCAGAUU .-----------......((((...(((((..........))).))...)))).((..((((((....)))))).))((((((((((((.((....))...))))))))))))... ( -29.50) >DroEre_CAF1 8037 111 + 1 UGAUUGCUAC---AUGUUUUAUAUGUGGA--AGGACUUACAUGACCCCAGUGGCGCUUACUCCCUAUGGGCAGUCGUUUGUAGCGCUUGACCAGAAGAACGCAGGCGUUGCAGAUU .(((((((((---((((...)))))))).--..............((((..((.(....).))...)))))))))((((((((((((((............)))))))))))))). ( -33.60) >DroYak_CAF1 8776 113 + 1 UCAUUGCUACUAUAUGGUUUAUAUGUGGA--A-UACUUACAUGACCCCAGUGACGCUUGCUACCGACAGGCAGUCGCUUGUAGCGCUUGACCAGAAGGACGCAGGCGUUGCAGAUU ..............(((..(.(((((((.--.-...))))))))..)))(((((((((((....).))))).)))))((((((((((((.((....))...))))))))))))... ( -35.70) >DroAna_CAF1 10729 109 + 1 UAAUUAGCA---AGUAUAUCACAUGAG----AGUACUUACGUGACCCCAGUGGCGUUUGCUGCCAGCCGGGAGGCGCUUGUAGCGUUUGACCAGCAAGACGCAGGCGUUGCAGAUC .........---......((((.((((----....)))).)))).....(..((((((((..(..((.((.((((((.....))))))..)).))..)..))))))))..)..... ( -34.10) >consensus U_A_________UUUGUUUUACAUAUGCA__AAUACUUACAUGACCCCAGUGACGCUUACUACCUAUGGGUAGUCGCUUGUAGCGCUUGACCAAAAGGACGCAGGCGUUGCAGAUU ................................................(((((((((((.......))))).))))))(((((((((((.((....))...))))))))))).... (-21.94 = -21.70 + -0.24)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:10 2006