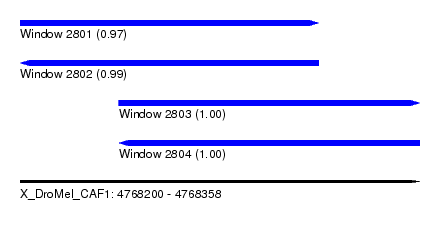
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,768,200 – 4,768,358 |
| Length | 158 |
| Max. P | 0.999972 |

| Location | 4,768,200 – 4,768,318 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 88.80 |
| Mean single sequence MFE | -49.33 |
| Consensus MFE | -44.51 |
| Energy contribution | -44.40 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4768200 118 + 22224390 CCAGUAUCCUGUAGUUGGACUCAAACAAGUCCCUC-CUUCCGCUCGACAGACGCUGGAAAAUCAAAGCCAGCGGAAGUGACUUCCGGCGGUUAAGAUUUUCCAGCGCCUGUGAAGCGCC .(((....))).....(((((......)))))...-....((((..((((.((((((((((((..((((..((((((...))))))..))))..)))))))))))).))))..)))).. ( -49.30) >DroSec_CAF1 6703 119 + 1 CCAGUGUCCUGUAGUUGGACUCCAACAAGUCCCUCCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGA .(((....))).....(((((......)))))......((((((..((((..((.((((..((..(((((.((((((...)))))).)))))..))..)))).))..))))..)))))) ( -49.30) >DroSim_CAF1 7305 119 + 1 CCAGCGUCCUGUAGUUGGACUCCAACAAGUCCCUCCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGA (((((........)))))....................((((((..((((..((.((((..((..(((((.((((((...)))))).)))))..))..)))).))..))))..)))))) ( -49.40) >consensus CCAGUGUCCUGUAGUUGGACUCCAACAAGUCCCUCCCUUCUGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGA .(((....))).....(((((......)))))......((((((..((((..((.((((..((..(((((.((((((...)))))).)))))..))..)))).))..))))..)))))) (-44.51 = -44.40 + -0.11)



| Location | 4,768,200 – 4,768,318 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 88.80 |
| Mean single sequence MFE | -55.87 |
| Consensus MFE | -52.30 |
| Energy contribution | -50.53 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.992042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4768200 118 - 22224390 GGCGCUUCACAGGCGCUGGAAAAUCUUAACCGCCGGAAGUCACUUCCGCUGGCUUUGAUUUUCCAGCGUCUGUCGAGCGGAAG-GAGGGACUUGUUUGAGUCCAACUACAGGAUACUGG ..(((((.(((((((((((((((((..(.(((.((((((...)))))).))).)..))))))))))))))))).)))))...(-(..((((((....))))))..)).(((....))). ( -60.70) >DroSec_CAF1 6703 119 - 1 UCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGGGACUUGUUGGAGUCCAACUACAGGACACUGG ((.((((.(((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).))))).)))).))..((..((((((....))))))..)).(((....))). ( -53.50) >DroSim_CAF1 7305 119 - 1 UCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGGGACUUGUUGGAGUCCAACUACAGGACGCUGG (((((((.(((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).))))).)))).....((..((((((....))))))..))...)))...... ( -53.40) >consensus UCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCAGAAGGGAGGGACUUGUUGGAGUCCAACUACAGGACACUGG ...((((.(((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).))))).)))).........((((((....)))))).....(((....))). (-52.30 = -50.53 + -1.77)



| Location | 4,768,239 – 4,768,358 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.07 |
| Mean single sequence MFE | -48.13 |
| Consensus MFE | -39.52 |
| Energy contribution | -39.67 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.82 |
| SVM decision value | 5.07 |
| SVM RNA-class probability | 0.999972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4768239 119 + 22224390 CGCUCGACAGACGCUGGAAAAUCAAAGCCAGCGGAAGUGACUUCCGGCGGUUAAGAUUUUCCAGCGCCUGUGAAGCGCCAGGAGCGAACUUGUUGGAGUCAAACUACAGGCUACCAGGG ((((..((((.((((((((((((..((((..((((((...))))))..))))..)))))))))))).))))..)))).((((......))))((((((((........)))).)))).. ( -51.40) >DroSec_CAF1 6743 119 + 1 UGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGAAGGAGCGAGAUUGUUGGAGUCAAACUUCAGGCUACCAGGG .((((.((((..((.((((..((..(((((.((((((...)))))).)))))..))..)))).))..))))....(....))))).......((((((((........)))).)))).. ( -49.30) >DroSim_CAF1 7345 93 + 1 UGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGAAGGAGCGAGCUUGU-------------------------- .(((((((((..((.((((..((..(((((.((((((...)))))).)))))..))..)))).))..))))....(....)...)))))....-------------------------- ( -43.70) >consensus UGCUCGACCGAAGCCGGAGCAUCGAAGCCGGCGGAAGUAGCUUCCGGCGGUUAAGAAACUCCAGCGCCGGUGAAGCGGAAGGAGCGAGCUUGUUGGAGUCAAACU_CAGGCUACCAGGG (((((.((((..((.((((..((..(((((.((((((...)))))).)))))..))..)))).))..))))....(....))))))......((((((((........)))).)))).. (-39.52 = -39.67 + 0.14)

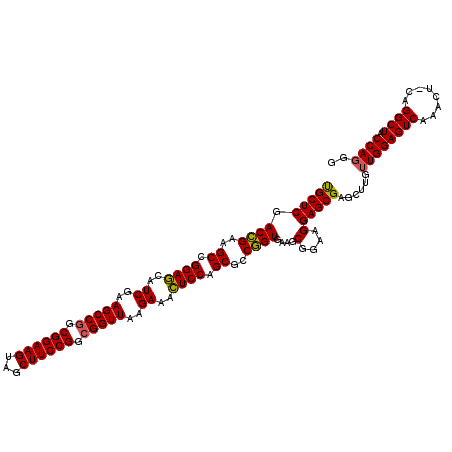
| Location | 4,768,239 – 4,768,358 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.07 |
| Mean single sequence MFE | -48.37 |
| Consensus MFE | -42.00 |
| Energy contribution | -40.23 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.17 |
| Mean z-score | -4.33 |
| Structure conservation index | 0.87 |
| SVM decision value | 4.59 |
| SVM RNA-class probability | 0.999925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4768239 119 - 22224390 CCCUGGUAGCCUGUAGUUUGACUCCAACAAGUUCGCUCCUGGCGCUUCACAGGCGCUGGAAAAUCUUAACCGCCGGAAGUCACUUCCGCUGGCUUUGAUUUUCCAGCGUCUGUCGAGCG ........(((.(.(((..((((......)))).))).).)))((((.(((((((((((((((((..(.(((.((((((...)))))).))).)..))))))))))))))))).)))). ( -55.40) >DroSec_CAF1 6743 119 - 1 CCCUGGUAGCCUGAAGUUUGACUCCAACAAUCUCGCUCCUUCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCA ....((.(((..((...(((....)))...))..)))))....((((.(((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).))))).)))). ( -47.60) >DroSim_CAF1 7345 93 - 1 --------------------------ACAAGCUCGCUCCUUCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCA --------------------------....((((((.......))...(((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).))))).)))). ( -42.10) >consensus CCCUGGUAGCCUG_AGUUUGACUCCAACAAGCUCGCUCCUUCCGCUUCACCGGCGCUGGAGUUUCUUAACCGCCGGAAGCUACUUCCGCCGGCUUCGAUGCUCCGGCUUCGGUCGAGCA ...........................................((((.(((((.((((((((.((..(.(((.((((((...)))))).))).)..)).)))))))).))))).)))). (-42.00 = -40.23 + -1.77)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:08 2006