| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,748,670 – 4,748,781 |
| Length | 111 |
| Max. P | 0.705026 |

| Location | 4,748,670 – 4,748,781 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 78.40 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -17.26 |
| Energy contribution | -18.68 |
| Covariance contribution | 1.43 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4748670 111 - 22224390 -GUC-UUUUCGCCAUUUU---UGCCAACGUUGUAAAGGGCACCAAAAAUAAUUAAGCUGGCAGAGGCAAUAAAAAUGCUUAACAAGCUUAAGGCCGGAAAUUCCCUAAGGAAGGUA -..(-((((((((.....---((((............))))..........(((((((.....(((((.......)))))....)))))))))).((.....))....)))))).. ( -25.90) >DroVir_CAF1 29687 102 - 1 -UCU-CUCCCGCU-UUACAGCCGCACAUAU---------UGGCAAAAAUAAUUAAGCGG-CUGAGGUAAUAAAAAUGCCUAACAAGCUGCU-CCAGGUAGACCGCAUAUGAACGUA -((.-....((.(-((((.((((.......---------))))...........(((((-((.(((((.......)))))....)))))))-....))))).)).....))..... ( -24.20) >DroSec_CAF1 41030 111 - 1 -GUC-UUUUCGCCAUUUU---UGCCCACGUUGUAAAGGGCACCAAAAAUAAUUAAGCUGGCAGAGGCAAUGAAAAUGCUUAACAAGCUUAAGGCCGGAAAUUCCCUAAGGAAGGUA -..(-((((((((.....---(((((..........)))))..........(((((((.....(((((.(....))))))....)))))))))).((.....))....)))))).. ( -29.70) >DroEre_CAF1 46935 112 - 1 -UUCUUUUUUGCCAUUUU---UCCCCACGAUGUAAAGGGCACCAGAAAUAAUUAAGCUGGCAGAGGCAAUAAAAAUGCUUAACAAGCUUAAGGCCGGAAAUUCCCUAUGGAAGGUA -........((((...((---(((...(........)(((...........(((((((.....(((((.......)))))....))))))).))))))))((((....)))))))) ( -24.30) >DroYak_CAF1 43720 111 - 1 -UUC-UUUUCGCCAUUUU---UGCCCACGUUGUAAAGGGCACCGAAAAUAAUUAAGCUGGCAGAGGCAAUAAAAAUGCUUAACAAGCUUAAGGCCGGAAACUCCCUAAGGAAGGUA -..(-((((((((.....---(((((..........)))))..........(((((((.....(((((.......)))))....)))))))))).(....).......)))))).. ( -29.90) >DroAna_CAF1 56516 111 - 1 UCCC-UUUUGGCUUUUUU---U-UUUUGGGGAAAAAGGGCACCAAAAAUAAUUAAGCUGGCAGAGGCAAUAAAAAUGCUUAACAAGCUUAAGGCCGGAAAUUCUCUAAGGAAGGUA ..((-((((((((.....---(-((((((............)))))))...(((((((.....(((((.......)))))....)))))))))))(((.....)))..)))))).. ( -27.40) >consensus _UUC_UUUUCGCCAUUUU___UGCCCACGUUGUAAAGGGCACCAAAAAUAAUUAAGCUGGCAGAGGCAAUAAAAAUGCUUAACAAGCUUAAGGCCGGAAAUUCCCUAAGGAAGGUA .....(((((((((((((...(((((..........)))))...)))))..(((((((.....(((((.......)))))....)))))))))).)))))((((....)))).... (-17.26 = -18.68 + 1.43)

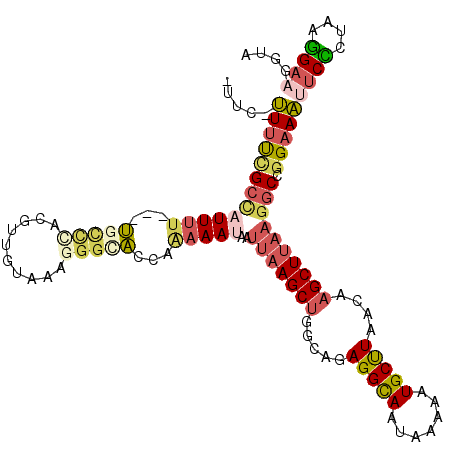

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:17:02 2006