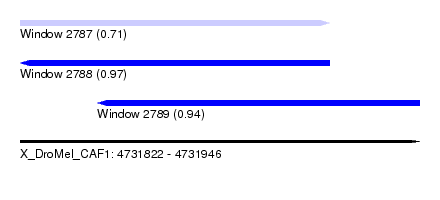
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,731,822 – 4,731,946 |
| Length | 124 |
| Max. P | 0.972333 |

| Location | 4,731,822 – 4,731,918 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 75.60 |
| Mean single sequence MFE | -25.89 |
| Consensus MFE | -3.08 |
| Energy contribution | -4.36 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.12 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4731822 96 + 22224390 ACACCAACGGACCAGAAGAAGUCGAAGGAAAUGUGCAACCUUUGGCU--G-CACAUAUUG-ACGAAGCUGAUAG----GAUGGCAAAUAUUCCGUACAUACGGA ...(((.(..(.(((.....(((((.....(((((((.((...)).)--)-))))).)))-))....))).)..----).))).......(((((....))))) ( -23.20) >DroSec_CAF1 24741 96 + 1 ACACCAACGGACCAGAAGAAGUCGAAGGAAAUGUGCAACCUUUGGCU--G-CACAUAUUG-ACGAAGCUGAUGG----GAUGGCAAAUAUUCCGCACAUGCGGA .......((..(((..((..(((((.....(((((((.((...)).)--)-))))).)))-))....))..)))----..))........(((((....))))) ( -29.30) >DroEre_CAF1 28155 97 + 1 ACACCAACGGACCAGAAGAAGUCGAAGGAAAUGUGCAACCUUAGGCU--G-CACAUAUUGGUCGAAGCAGAUGG----GAUGGCAAAUAUUUCGCUCGUAGGGA ...((.(((((((((......((....)).(((((((.((...)).)--)-))))).)))))).....((.((.----.(((.....)))..))))))).)).. ( -28.10) >DroYak_CAF1 25974 96 + 1 ACACCAACGGACCAGAAGAAGUCGAAGGAAAUGUGCAACCCUAGGUU--G-CACAUAUUG-ACGAAGCAGAUGG----GAUGGCAAAUAUGCCGCACAUGGGGC ...(((.....(((......(((((.....((((((((((...))))--)-))))).)))-))........)))----(.(((((....))))))...)))... ( -28.94) >DroAna_CAF1 27824 95 + 1 ACAGCAUCCGAACAGAACCAGAAGCUGGAAAAGUGCAAACCAAGAUGUAGCCAGAACGUG-GCUCAGCUGACACGUAAGAU-CCGAAUACCCUGCAC------- .((((.((.(........).)).)))).....(((((.....((.((.(((((.....))-))))).))....(....)..-..........)))))------- ( -19.90) >consensus ACACCAACGGACCAGAAGAAGUCGAAGGAAAUGUGCAACCUUAGGCU__G_CACAUAUUG_ACGAAGCUGAUGG____GAUGGCAAAUAUUCCGCACAUACGGA ..............................((((((........(((........((((..........))))........))).........))))))..... ( -3.08 = -4.36 + 1.28)


| Location | 4,731,822 – 4,731,918 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 75.60 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -8.16 |
| Energy contribution | -7.28 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.29 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4731822 96 - 22224390 UCCGUAUGUACGGAAUAUUUGCCAUC----CUAUCAGCUUCGU-CAAUAUGUG-C--AGCCAAAGGUUGCACAUUUCCUUCGACUUCUUCUGGUCCGUUGGUGU (((((....)))))......((((.(----..(((((..(((.-....(((((-(--((((...))))))))))......)))......)))))..).)))).. ( -29.10) >DroSec_CAF1 24741 96 - 1 UCCGCAUGUGCGGAAUAUUUGCCAUC----CCAUCAGCUUCGU-CAAUAUGUG-C--AGCCAAAGGUUGCACAUUUCCUUCGACUUCUUCUGGUCCGUUGGUGU (((((....)))))............----.(((((((.....-....(((((-(--((((...)))))))))).......((((......)))).))))))). ( -32.70) >DroEre_CAF1 28155 97 - 1 UCCCUACGAGCGAAAUAUUUGCCAUC----CCAUCUGCUUCGACCAAUAUGUG-C--AGCCUAAGGUUGCACAUUUCCUUCGACUUCUUCUGGUCCGUUGGUGU ......((((((..............----.....))).)))(((((((((((-(--((((...)))))))))).......((((......)))).)))))).. ( -25.91) >DroYak_CAF1 25974 96 - 1 GCCCCAUGUGCGGCAUAUUUGCCAUC----CCAUCUGCUUCGU-CAAUAUGUG-C--AACCUAGGGUUGCACAUUUCCUUCGACUUCUUCUGGUCCGUUGGUGU ((((.(((..(((((....))))...----.........(((.-....(((((-(--((((...))))))))))......))).........)..))).)).)) ( -28.10) >DroAna_CAF1 27824 95 - 1 -------GUGCAGGGUAUUCGG-AUCUUACGUGUCAGCUGAGC-CACGUUCUGGCUACAUCUUGGUUUGCACUUUUCCAGCUUCUGGUUCUGUUCGGAUGCUGU -------((((((((((..(((-(....(((((.(......).-)))))))))..))).......))))))).....((((.(((((......))))).)))). ( -26.81) >consensus UCCCCAUGUGCGGAAUAUUUGCCAUC____CCAUCAGCUUCGU_CAAUAUGUG_C__AGCCUAAGGUUGCACAUUUCCUUCGACUUCUUCUGGUCCGUUGGUGU ...................(((((......(((......(((......(((((....((((...)))).)))))......))).......))).....))))). ( -8.16 = -7.28 + -0.88)



| Location | 4,731,846 – 4,731,946 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 82.29 |
| Mean single sequence MFE | -28.73 |
| Consensus MFE | -17.35 |
| Energy contribution | -17.10 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4731846 100 - 22224390 AUCUCCAAGUUGCCGA------UGCUACCACGCAUCCGUAUGUACGGAAUAUUUGCCAUCCUAUCAGCUUCGU-CAAUAUGUGCAGCCAAAGGUUGCACAUUUCCUU ........((((.(((------.(((.....((((((((....))))).....))).........))).))).-))))((((((((((...))))))))))...... ( -27.34) >DroSec_CAF1 24765 100 - 1 AUGUCCAAGUCGCCGU------UGCUACCACGCAUCCGCAUGUGCGGAAUAUUUGCCAUCCCAUCAGCUUCGU-CAAUAUGUGCAGCCAAAGGUUGCACAUUUCCUU ........(.((..((------((.......((((((((....))))).....)))........))))..)).-)...((((((((((...))))))))))...... ( -27.26) >DroEre_CAF1 28179 101 - 1 UACUGCACGUCGCCGA------CGAUACCUCGCAUCCCUACGAGCGAAAUAUUUGCCAUCCCAUCUGCUUCGACCAAUAUGUGCAGCCUAAGGUUGCACAUUUCCUU ....(((((((...))------)).....((((.((.....))))))..................)))..........((((((((((...))))))))))...... ( -23.20) >DroYak_CAF1 25998 106 - 1 UACUCAAAGUCGCCGACGACGACGAUGCCACACAGCCCCAUGUGCGGCAUAUUUGCCAUCCCAUCUGCUUCGU-CAAUAUGUGCAACCUAGGGUUGCACAUUUCCUU ........((((....))))(((((.((..((((......)))).((((....)))).........)).))))-)...((((((((((...))))))))))...... ( -37.10) >consensus AACUCCAAGUCGCCGA______CGAUACCACGCAUCCCCAUGUGCGGAAUAUUUGCCAUCCCAUCAGCUUCGU_CAAUAUGUGCAGCCAAAGGUUGCACAUUUCCUU ..................................(((((....)))))..............................((((((((((...))))))))))...... (-17.35 = -17.10 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:54 2006