
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,719,100 – 4,719,238 |
| Length | 138 |
| Max. P | 0.921220 |

| Location | 4,719,100 – 4,719,198 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 66.12 |
| Mean single sequence MFE | -24.78 |
| Consensus MFE | -15.81 |
| Energy contribution | -15.93 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4719100 98 - 22224390 CAGAUGUGGUGGCUAAGGGGGCGUGGCAGGGUCAGACAAGAUGUUGGCAACAAAAGCCAAAAGUGCCAUCGCG-------UAAAAAUAAAACAACACGACGGCUC ...((((((((((......(((.((.(((.(((......))).))).))......)))......)))))))))-------)........................ ( -28.80) >DroSec_CAF1 11962 94 - 1 UAA----GGGGGCUAAGGGGGCGUGGCAGGGUCAGACUGGAUGUUGGCAACAAAAGCCAAAAGUGCCACCGCG-------UAAAAAUAAAACAACAGGACGGCCC ...----..(((((......((((((...(((.....(((...(((....)))...))).....))).)))))-------)..........(....)...))))) ( -25.90) >DroMoj_CAF1 16903 99 - 1 AAU----GGGGGUAGAUCGGGUCUGGUCUGGUAAAA-GCAACAUUGGCAACAAAAGCUAAAAGUGCCUCAGCGAGUGGAGUAAGAAAAAAAUAUUUGUGCAGUG- ..(----((((.(((((((....))))))).....(-((....(((....)))..))).......)))))(((((((..............)))))))......- ( -19.64) >consensus AAA____GGGGGCUAAGGGGGCGUGGCAGGGUCAGACAAGAUGUUGGCAACAAAAGCCAAAAGUGCCACCGCG_______UAAAAAUAAAACAACAGGACGGCCC .......((((((......(((.((.(((.(((......))).))).))......)))......))))))................................... (-15.81 = -15.93 + 0.13)

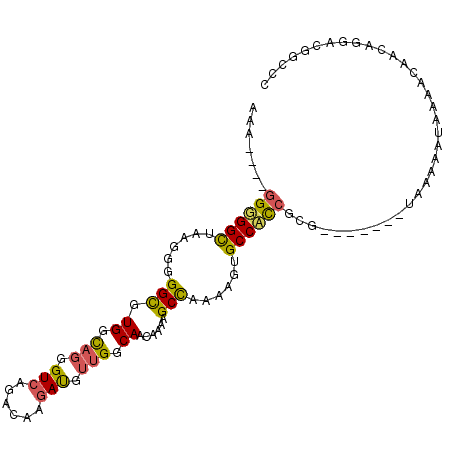
| Location | 4,719,125 – 4,719,238 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.78 |
| Mean single sequence MFE | -33.69 |
| Consensus MFE | -20.96 |
| Energy contribution | -22.28 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.907982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4719125 113 - 22224390 UCAAAGUUGAGUUCGCUUAAGAGGCGAACGGCAAGGGGGCCAGAUGUGGUGGCUAAGGGGGCGUGGCAGGGUCAGACAAGAUGUUGGCAACAAAAGCCAAAAGUGCCAUCGCG------- ..........(((((((.....)))))))(((......)))....((((((((......(((.((.(((.(((......))).))).))......)))......)))))))).------- ( -42.10) >DroSec_CAF1 11987 109 - 1 UCAAAGUUGAGUUCUCUUAAGAGGCGGACGGCAAGGGGGCUAA----GGGGGCUAAGGGGGCGUGGCAGGGUCAGACUGGAUGUUGGCAACAAAAGCCAAAAGUGCCACCGCG------- .............(((....)))((((..((((....((((..----.(..(((((.(.((..((((...))))..))...).)))))..)...)))).....)))).)))).------- ( -32.90) >DroEre_CAF1 14533 108 - 1 UCAAAGUUGAGUUCGCUUAAGAGGCGGACGGCAAGGGG-CUAGA---AAGGGCUAA-GGGGCGUGGCAGGGUCAGACAGGAUGUUGGCAACAAAAGCCAAAAGUGCCACCGCG------- ......(((.(((((((.....)))))))..)))((((-((...---...))))..-((.((.((((...(((......))).(((....)))..))))...)).)).))...------- ( -33.60) >DroYak_CAF1 12469 93 - 1 UCAAAGUUGAGUUCGCCUAUGAGGCGGACGGCAAAG-------------------A-GGGGCGUGGCAGGGUCAGACAGGAUGUUGGCAACAAAAGCCAAAAGUGCCACCGCG------- ((....(((.(((((((.....)))))))..)))..-------------------.-.))((((((((..(((......))).(((((.......)))))...))))).))).------- ( -32.70) >DroMoj_CAF1 16927 111 - 1 CCAAAGCAAAGGCAAUC---GAACAGAACGGAAGGG-AGGAAU----GGGGGUAGAUCGGGUCUGGUCUGGUAAAA-GCAACAUUGGCAACAAAAGCUAAAAGUGCCUCAGCGAGUGGAG (((..((..(((((...---...((((.((((...(-(.....----.........))...)))).)))).....(-((....(((....)))..))).....)))))..))...))).. ( -27.14) >consensus UCAAAGUUGAGUUCGCUUAAGAGGCGGACGGCAAGGGGGCUAG____GGGGGCUAA_GGGGCGUGGCAGGGUCAGACAGGAUGUUGGCAACAAAAGCCAAAAGUGCCACCGCG_______ ..........(((((((.....)))))))...............................((((((((..(((......))).(((((.......)))))...))))).)))........ (-20.96 = -22.28 + 1.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:50 2006