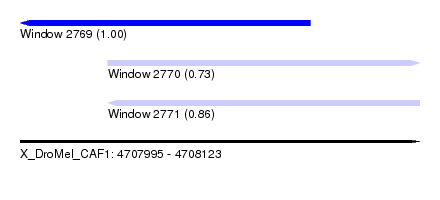
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,707,995 – 4,708,123 |
| Length | 128 |
| Max. P | 0.996642 |

| Location | 4,707,995 – 4,708,088 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.60 |
| Mean single sequence MFE | -30.00 |
| Consensus MFE | -19.64 |
| Energy contribution | -20.87 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4707995 93 - 22224390 UCAAUAUGCCUUGAAAGGGUUGCGGGUUGCGG-----------AAAAG----CAGCUGCGGUGGCUUAAGUGAGGCAUAAGAAAAUAAUUAAAAACAGAAAG------------CGAUUU ....(((((((((...((((..(.((((((..-----------....)----)))))...)..))))...))))))))).......................------------...... ( -25.70) >DroPse_CAF1 21092 119 - 1 UCAAUAUGCCAC-AAAGGGUUGCAGAGAGGGGCUGGUGGCUGGUGCGGCCGUUGGCUGUGGUGGCUUAAGUGAGGCAUAAGAAAAUAAUUAAAAACACAAAAGAGCCGAACACACAGUUU ....((((((.(-(..((((..(.....(.((((..((((((...))))))..)))).).)..))))...)).))))))............................((((.....)))) ( -32.60) >DroPer_CAF1 20512 116 - 1 UCAAUAUGCCAC-AAAGGGUUGCAGAGAGGGGCC-AUGGCUGGUGAGGCCGUUGGCUGUGGUGGCUUAAGUGAGGCAUAAGAAAAUAAUUAAAAACACAAAAGAGCCGAAC--ACAGUUU ....((((((.(-(..((((..(.....(.((((-((((((.....))))).))))).).)..))))...)).))))))................................--....... ( -31.70) >consensus UCAAUAUGCCAC_AAAGGGUUGCAGAGAGGGGC___UGGCUGGUGAGGCCGUUGGCUGUGGUGGCUUAAGUGAGGCAUAAGAAAAUAAUUAAAAACACAAAAGAGCCGAAC__ACAGUUU ....((((((......((((..(.....(.((((.((((((.....)))))).)))).).)..))))......))))))......................................... (-19.64 = -20.87 + 1.23)

| Location | 4,708,023 – 4,708,123 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.65 |
| Mean single sequence MFE | -25.47 |
| Consensus MFE | -16.90 |
| Energy contribution | -16.73 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4708023 100 + 22224390 UUAUGCCUCACUUAAGCCACCGCAGCUG----CUUUU-----------CCGCAACCCGCAACCCUUUCAAGGCAUAUUGAGUGCUUUGUAAUUUUUGCGGUUUUUAAGCCUUUUC----- ..........((((((..((((((((((----(....-----------..)))....))........((((((((.....)))))))).......)))))).)))))).......----- ( -23.20) >DroPse_CAF1 21132 119 + 1 UUAUGCCUCACUUAAGCCACCACAGCCAACGGCCGCACCAGCCACCAGCCCCUCUCUGCAACCCUUU-GUGGCAUAUUGAGUGCUUUGUAAUUUUUGUGGGUCUUAAGCCUUUCCACCAC ..........((((((.(.((((((((...))).((((((((((((((.......))).........-)))))....)).))))...........)))))).))))))............ ( -26.60) >DroPer_CAF1 20550 118 + 1 UUAUGCCUCACUUAAGCCACCACAGCCAACGGCCUCACCAGCCAU-GGCCCCUCUCUGCAACCCUUU-GUGGCAUAUUGAGUGCUUUGUAAUUUUUGUGGGUCUUAAGCCUUUCCACCAC ..........((((((.(.(((((((((..(((.......))).)-))).......(((((......-..(((((.....)))))))))).....)))))).))))))............ ( -26.60) >consensus UUAUGCCUCACUUAAGCCACCACAGCCAACGGCCUCACCAGCCA___GCCCCUCUCUGCAACCCUUU_GUGGCAUAUUGAGUGCUUUGUAAUUUUUGUGGGUCUUAAGCCUUUCCACCAC ..........((((((...((((((.....(((.......)))..............((((.........(((((.....))))))))).....))))))..))))))............ (-16.90 = -16.73 + -0.16)


| Location | 4,708,023 – 4,708,123 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.65 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -22.87 |
| Energy contribution | -24.21 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4708023 100 - 22224390 -----GAAAAGGCUUAAAAACCGCAAAAAUUACAAAGCACUCAAUAUGCCUUGAAAGGGUUGCGGGUUGCGG-----------AAAAG----CAGCUGCGGUGGCUUAAGUGAGGCAUAA -----......((((...................))))......(((((((((...((((..(.((((((..-----------....)----)))))...)..))))...))))))))). ( -27.11) >DroPse_CAF1 21132 119 - 1 GUGGUGGAAAGGCUUAAGACCCACAAAAAUUACAAAGCACUCAAUAUGCCAC-AAAGGGUUGCAGAGAGGGGCUGGUGGCUGGUGCGGCCGUUGGCUGUGGUGGCUUAAGUGAGGCAUAA ((((.(.............)))))....................((((((.(-(..((((..(.....(.((((..((((((...))))))..)))).).)..))))...)).)))))). ( -36.92) >DroPer_CAF1 20550 118 - 1 GUGGUGGAAAGGCUUAAGACCCACAAAAAUUACAAAGCACUCAAUAUGCCAC-AAAGGGUUGCAGAGAGGGGCC-AUGGCUGGUGAGGCCGUUGGCUGUGGUGGCUUAAGUGAGGCAUAA ((((.(.............)))))....................((((((.(-(..((((..(.....(.((((-((((((.....))))).))))).).)..))))...)).)))))). ( -36.12) >consensus GUGGUGGAAAGGCUUAAGACCCACAAAAAUUACAAAGCACUCAAUAUGCCAC_AAAGGGUUGCAGAGAGGGGC___UGGCUGGUGAGGCCGUUGGCUGUGGUGGCUUAAGUGAGGCAUAA ...........(((((((..((((............((((((..............))).))).......((((.((((((.....)))))).))))))))...)))))))......... (-22.87 = -24.21 + 1.34)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:38 2006