
| Sequence ID | X_DroMel_CAF1 |
|---|---|
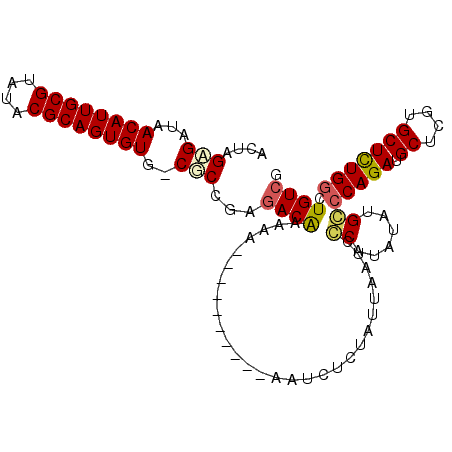
| Location | 4,704,415 – 4,704,532 |
| Length | 117 |
| Max. P | 0.778018 |

| Location | 4,704,415 – 4,704,515 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 82.59 |
| Mean single sequence MFE | -27.81 |
| Consensus MFE | -20.81 |
| Energy contribution | -21.25 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4704415 100 - 22224390 GCUAGCGAUAACAUUGCGUAUACGCAGUGUU-CGCCGAGACAAAAAAAAAAA----AAACUGUAUUAAUCCCAUAUAUGGGCAGAUGCUCGUGCUCUGGCUGUCG ....((((..((((((((....)))))))))-)))...((((..........----.............((((....))))((((.((....))))))..)))). ( -30.40) >DroSec_CAF1 57303 105 - 1 ACUAGAGAUAACAUUGCGUAUACGCAGUGUUGCGCAGAGACAAAAAAAAAAAAGGAAAUCUCUAUUAAUCCCAUAUAUGCCCAGAUGCUCGUGCUCUGGCUGUCG .((.(.(.((((((((((....))))))))))).))).((((...........(((............))).......(((.(((.((....)))))))))))). ( -26.70) >DroEre_CAF1 58791 94 - 1 GCUCGAGAUAACAUUGCGUAUACGCAGUGUG-CUCCGAUACAAAAA----------GAUCUCCAUUAAGCGCAUAUAUGCCCAGAUGCUCGUGCUUUGGCUGUCG ..(((((...((((((((....)))))))).-)).)))........----------(((..(((..((((((......((......))..)))))))))..))). ( -26.20) >DroYak_CAF1 55627 94 - 1 ACUCGAGACAACAUUGCGUAUACGCAGUGUG-CGCUGAGACGAAAA----------AAUCUCUAUUAAUCGCAUAUGCGCCCAGAUGCUCGUGCUUUGGCUGUCG ......((((((((((((....))))))))(-(((.((((......----------..))))..............))))(((((.((....))))))).)))). ( -27.96) >consensus ACUAGAGAUAACAUUGCGUAUACGCAGUGUG_CGCCGAGACAAAAA__________AAUCUCUAUUAAUCCCAUAUAUGCCCAGAUGCUCGUGCUCUGGCUGUCG ....(((...((((((((....))))))))..)))...((((............................((......))(((((.((....))))))).)))). (-20.81 = -21.25 + 0.44)


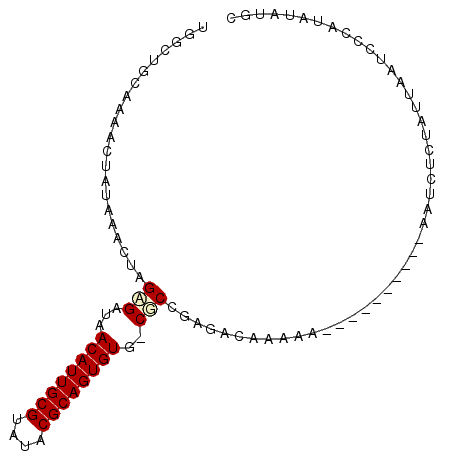
| Location | 4,704,440 – 4,704,532 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 76.83 |
| Mean single sequence MFE | -19.25 |
| Consensus MFE | -11.90 |
| Energy contribution | -12.28 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4704440 92 - 22224390 UGGUUGCAAAGCUAUAAGCUAGCGAUAACAUUGCGUAUACGCAGUGUU-CGCCGAGACAAAAAAAAAAA----AAACUGUAUUAAUCCCAUAUAUGG .((.((((.(((.....))).((((..((((((((....)))))))))-))).................----....)))).....))......... ( -20.20) >DroSec_CAF1 57328 97 - 1 UGACUGCAAAGCUAUAAACUAGAGAUAACAUUGCGUAUACGCAGUGUUGCGCAGAGACAAAAAAAAAAAAGGAAAUCUCUAUUAAUCCCAUAUAUGC ...((((....(((.....))).(.((((((((((....)))))))))))))))................(((............)))......... ( -19.30) >DroEre_CAF1 58816 86 - 1 UGGCUACACAACUAUAAGCUCGAGAUAACAUUGCGUAUACGCAGUGUG-CUCCGAUACAAAAA----------GAUCUCCAUUAAGCGCAUAUAUGC ..(((................(((...((((((((....)))))))).-))).(((.......----------.))).......)))(((....))) ( -18.50) >DroYak_CAF1 55652 80 - 1 UG------AAACUAUAAACUCGAGACAACAUUGCGUAUACGCAGUGUG-CGCUGAGACGAAAA----------AAUCUCUAUUAAUCGCAUAUGCGC ..------..........(((.((...((((((((....)))))))).-..))))).......----------.............(((....))). ( -19.00) >consensus UGGCUGCAAAACUAUAAACUAGAGAUAACAUUGCGUAUACGCAGUGUG_CGCCGAGACAAAAA__________AAUCUCUAUUAAUCCCAUAUAUGC .....................(((...((((((((....))))))))..)))............................................. (-11.90 = -12.28 + 0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:34 2006