
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,703,595 – 4,703,782 |
| Length | 187 |
| Max. P | 0.995910 |

| Location | 4,703,595 – 4,703,702 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 74.69 |
| Mean single sequence MFE | -29.31 |
| Consensus MFE | -18.64 |
| Energy contribution | -18.70 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.995910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
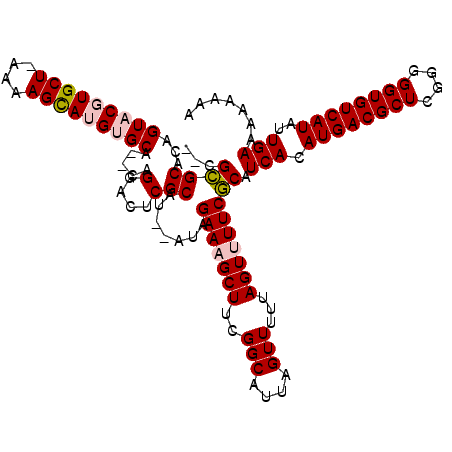
>X_DroMel_CAF1 4703595 107 + 22224390 A-GCCACACAAAUUUG--UUUUUUAUUUUUUUUAACACACUUAUACGCCCCACUUUUCUCGCGGUGUGUCGUCUGAGCGCACAGUACGUGCUAAAAGUAUGUGCACAGGA .-.((...........--...................(.((((.(((.(.((((........)))).).))).)))).)....((((((((.....))))))))...)). ( -20.50) >DroSec_CAF1 56504 101 + 1 A-GCCGCACAUAUUUAACAUUUUUUUUUUUUUUUACACACUUAUAC-CCUCGCUUUUCUCUCGGUGUG-------CGCGCACAGUACGUGCUAAAAGUAUGUGCACAGGA .-...((((((((((....................((((((.....-...............))))))-------...((((.....))))...))))))))))...... ( -22.55) >DroEre_CAF1 57979 101 + 1 AUGCCACACAUCUGGCACAU------UGUUUUUUGCACACUUGA---CCCUUCUUUUCGCACUGUGUGUCGUCUGCGCGCACAGUACGUGCUAAAAGCAUGUGCACAGGA ..........((((((((((------.((((((.((((....((---....)).....(.((((((((.((....)))))))))).))))).)))))))))))).)))). ( -37.10) >DroYak_CAF1 54776 101 + 1 AUGCCACACACUUUUCACAU------AGUUGGGUACACACUUGC---CCCUACUUUUCUCUCAGUGUGUGGCGAGCGCGCACAGUACGUGCUAAAAGUAUGUGCACAGGA .(((((((((((........------(((.(((((......)))---))..)))........)))))))))))..........((((((((.....))))))))...... ( -37.09) >consensus A_GCCACACAUAUUUCACAU______UGUUUUUUACACACUUAU___CCCCACUUUUCUCUCGGUGUGUCGUCUGCGCGCACAGUACGUGCUAAAAGUAUGUGCACAGGA ...((..........................................................((((((.......)))))).((((((((.....))))))))...)). (-18.64 = -18.70 + 0.06)


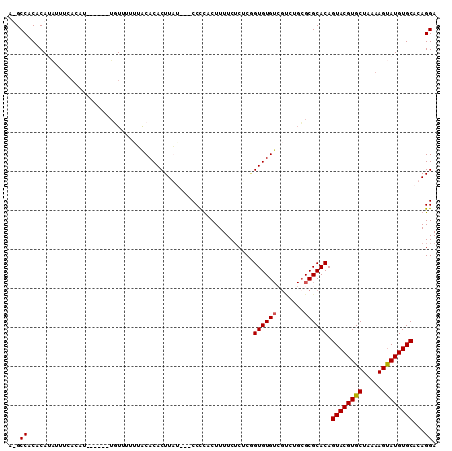
| Location | 4,703,595 – 4,703,702 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 74.69 |
| Mean single sequence MFE | -30.82 |
| Consensus MFE | -15.16 |
| Energy contribution | -14.98 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4703595 107 - 22224390 UCCUGUGCACAUACUUUUAGCACGUACUGUGCGCUCAGACGACACACCGCGAGAAAAGUGGGGCGUAUAAGUGUGUUAAAAAAAAUAAAAAA--CAAAUUUGUGUGGC-U ......((.(((((((((((((((...((((((((..(......).((((.......))))))))))))..)))))))))).((((......--...)))))))))))-. ( -26.50) >DroSec_CAF1 56504 101 - 1 UCCUGUGCACAUACUUUUAGCACGUACUGUGCGCG-------CACACCGAGAGAAAAGCGAGG-GUAUAAGUGUGUAAAAAAAAAAAAAAAUGUUAAAUAUGUGCGGC-U .....((((((((......((((.....))))(((-------(((..(....)....((....-))....))))))......................))))))))..-. ( -22.40) >DroEre_CAF1 57979 101 - 1 UCCUGUGCACAUGCUUUUAGCACGUACUGUGCGCGCAGACGACACACAGUGCGAAAAGAAGGG---UCAAGUGUGCAAAAAACA------AUGUGCCAGAUGUGUGGCAU ...((..(((.(((((((....(((((((((.(((....)).).))))))))).....)))))---.)).)))..)).......------..((((((......)))))) ( -35.20) >DroYak_CAF1 54776 101 - 1 UCCUGUGCACAUACUUUUAGCACGUACUGUGCGCGCUCGCCACACACUGAGAGAAAAGUAGGG---GCAAGUGUGUACCCAACU------AUGUGAAAAGUGUGUGGCAU ....(((((((((((((...((((((..(((((((((.(((.(..(((........)))..))---)).))))))))).....)------))))).))))))))).)))) ( -39.20) >consensus UCCUGUGCACAUACUUUUAGCACGUACUGUGCGCGCAGACGACACACCGAGAGAAAAGUAGGG___ACAAGUGUGUAAAAAAAA______AUGUGAAAUAUGUGUGGC_U .((.((((((((((.........))).)))))))........(((((.......................)))))..............................))... (-15.16 = -14.98 + -0.19)

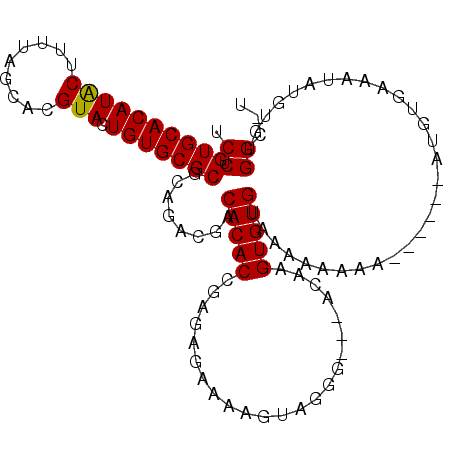
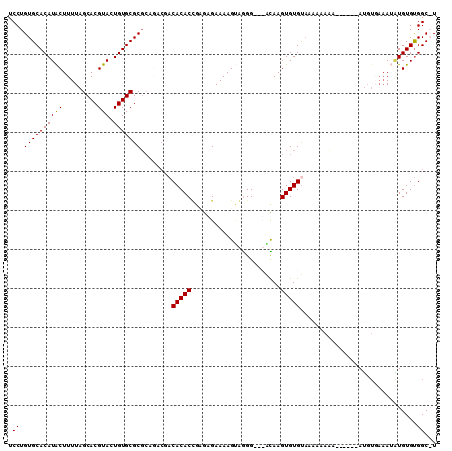
| Location | 4,703,665 – 4,703,782 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.47 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -30.34 |
| Energy contribution | -31.15 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
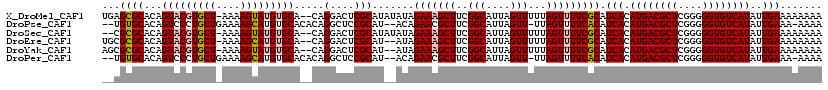
>X_DroMel_CAF1 4703665 117 + 22224390 UGAGCGCACAGUACGUGCU-AAAAGUAUGUGCA--CAGGACUCGCAUAUAUAGAAAGCUUCGGCAUUAGUUUUUAGUUUUCGCAUCACAUGACGCUCGGGGGUGUCAUAUUGAAAAAAAA (((((((((.....)))).-....((((((((.--........)))))))).(((((((..(((....)))...))))))))).))).((((((((....))))))))............ ( -33.20) >DroPse_CAF1 16826 114 + 1 --UGUGCACAGUCCCUGCUGAAAAGCAUGUGCACACAGGCUCCGCAU--ACAGAACGCUUCGGCAUUAGUU-UUAGUUUUCACAUCACAUGACGCUCGGGGGUGUCAUAUUGAAA-AAAA --((((((((.....((((....))))))))))))............--.......((....)).......-....((((((......((((((((....))))))))..)))))-)... ( -32.60) >DroSec_CAF1 56570 115 + 1 --CGCGCACAGUACGUGCU-AAAAGUAUGUGCA--CAGGACUCGCAUAUAUAGAAAGCUUCGGCAUUAGUUUUUAGUUUUCGCAUCACAUGACGCUCGGGGGUGUCAUAUUGAAAAAAAA --.((((((.....)))).-....((((((((.--........)))))))).(((((((..(((....)))...))))))))).(((.((((((((....))))))))..)))....... ( -33.30) >DroEre_CAF1 58043 115 + 1 UGCGCGCACAGUACGUGCU-AAAAGCAUGUGCA--CAGGACUCGCAU--AUAGAAAGCUUCGGCAUUAGUUUUUAGUUUUCGCAUCACAUGACGCUCGGGGGUGUCAUAUUGAAAAAAAA ((((((..(.((((((((.-....)))))))).--...)...)))..--...(((((((..(((....)))...))))))))))(((.((((((((....))))))))..)))....... ( -34.90) >DroYak_CAF1 54840 115 + 1 AGCGCGCACAGUACGUGCU-AAAAGUAUGUGCA--CAGGACUCGCAU--AUAGAAAGCUUCGGCAUUAGUUUUUAGUUUUCGCAUCACAUGACGCUCGGGGGUGUCAUAUUGAAAAAAAA .(((((..(.((((((((.-....)))))))).--...)...)))..--...(((((((..(((....)))...))))))))).(((.((((((((....))))))))..)))....... ( -32.60) >DroPer_CAF1 16185 114 + 1 --UGUGCACAGUCCCUGCUGAAAAGCAUGUGCACACAGGCUCCGCAU--ACAGAACGCUUCGGCAUUAGUU-UUAGUUUUCACAUCACAUGACGCUCGGGGGUGUCAUAUUGAAA-AAAA --((((((((.....((((....))))))))))))............--.......((....)).......-....((((((......((((((((....))))))))..)))))-)... ( -32.60) >consensus __CGCGCACAGUACGUGCU_AAAAGCAUGUGCA__CAGGACUCGCAU__AUAGAAAGCUUCGGCAUUAGUUUUUAGUUUUCGCAUCACAUGACGCUCGGGGGUGUCAUAUUGAAAAAAAA ...((((...(((((((((....))))))))).....(....))).......(((((((..(((....)))...))))))))).(((.((((((((....))))))))..)))....... (-30.34 = -31.15 + 0.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:33 2006