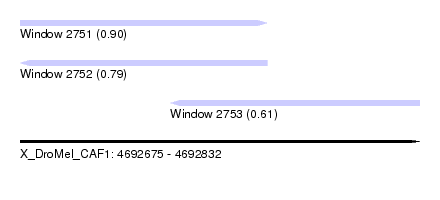
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,692,675 – 4,692,832 |
| Length | 157 |
| Max. P | 0.897605 |

| Location | 4,692,675 – 4,692,772 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 88.35 |
| Mean single sequence MFE | -28.42 |
| Consensus MFE | -21.60 |
| Energy contribution | -21.36 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4692675 97 + 22224390 ACAGGGCACAGAACGCACAGCCAUGAGACAUGGCCUGCCAAGAUAUAGGCACA----CACAAUUUGCCUCGACUGUCGAGUGCGAUAAAUUUGGGCUUAUG ....((((..(....)...((((((...)))))).))))....(((((((.((----......((((((((.....)))).))))......)).))))))) ( -27.50) >DroSec_CAF1 46101 90 + 1 ACGGGGUAAAA-------GGCCAUGAGACAUGGCCUGCCAAGAUAUAGGCACA----CACAAUUUGCCUCGACUGUCGAGUGCGAUAAAUUUGGGCUUAUG .((((((((((-------(((((((...))))))))(((........)))...----.....)))))))))......((((.(((.....))).))))... ( -34.30) >DroSim_CAF1 19211 90 + 1 ACGGGGUAAAA-------GGCCAUGAGACAUGGCCUGCCAAGAUAUAGGCACA----CACAAUUUGCCUCGACUGUCGAGUGCGAUAAAUUUGGGCUUAUG .((((((((((-------(((((((...))))))))(((........)))...----.....)))))))))......((((.(((.....))).))))... ( -34.30) >DroEre_CAF1 46531 83 + 1 -------ACAC-------AGCCAUGAGACAUGGCCUGCCAAGAUAUAGGCACA----CACAAUUUGCCUCGACUGUCGAGUGCGAUAAAUUUGGGCUUAUG -------...(-------(((((((...))))).)))......(((((((.((----......((((((((.....)))).))))......)).))))))) ( -24.20) >DroYak_CAF1 43458 94 + 1 ACACAGCACAC-------AGCCAAGAGACAUGGCCUGCCAAGAUAUAGGCACACACACACAAUUUGCCUCGACUGUCGAGUGCGAUAAAUUUGGGCUUAUG ...........-------((((........(((((((........))))).))......(((.((((((((.....)))).)))).....))))))).... ( -21.80) >consensus ACAGGGCACAA_______AGCCAUGAGACAUGGCCUGCCAAGAUAUAGGCACA____CACAAUUUGCCUCGACUGUCGAGUGCGAUAAAUUUGGGCUUAUG ..................((((..........(((((........))))).........(((.((((((((.....)))).)))).....))))))).... (-21.60 = -21.36 + -0.24)



| Location | 4,692,675 – 4,692,772 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 88.35 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -22.38 |
| Energy contribution | -22.34 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4692675 97 - 22224390 CAUAAGCCCAAAUUUAUCGCACUCGACAGUCGAGGCAAAUUGUG----UGUGCCUAUAUCUUGGCAGGCCAUGUCUCAUGGCUGUGCGUUCUGUGCCCUGU ..................((((.((((((((((((((...((.(----(.((((........)))).))))))))))..)))))).))....))))..... ( -31.60) >DroSec_CAF1 46101 90 - 1 CAUAAGCCCAAAUUUAUCGCACUCGACAGUCGAGGCAAAUUGUG----UGUGCCUAUAUCUUGGCAGGCCAUGUCUCAUGGCC-------UUUUACCCCGU ..................((.((((.....))))))........----..((((........))))(((((((...)))))))-------........... ( -25.00) >DroSim_CAF1 19211 90 - 1 CAUAAGCCCAAAUUUAUCGCACUCGACAGUCGAGGCAAAUUGUG----UGUGCCUAUAUCUUGGCAGGCCAUGUCUCAUGGCC-------UUUUACCCCGU ..................((.((((.....))))))........----..((((........))))(((((((...)))))))-------........... ( -25.00) >DroEre_CAF1 46531 83 - 1 CAUAAGCCCAAAUUUAUCGCACUCGACAGUCGAGGCAAAUUGUG----UGUGCCUAUAUCUUGGCAGGCCAUGUCUCAUGGCU-------GUGU------- ((((.(((..........((.((((.....))))))....((.(----(.((((........)))).))))........))))-------))).------- ( -24.60) >DroYak_CAF1 43458 94 - 1 CAUAAGCCCAAAUUUAUCGCACUCGACAGUCGAGGCAAAUUGUGUGUGUGUGCCUAUAUCUUGGCAGGCCAUGUCUCUUGGCU-------GUGUGCUGUGU ..............(((.((((...(((((((((((.....))(((.((.((((........)))).)))))....)))))))-------)))))).))). ( -28.40) >consensus CAUAAGCCCAAAUUUAUCGCACUCGACAGUCGAGGCAAAUUGUG____UGUGCCUAUAUCUUGGCAGGCCAUGUCUCAUGGCU_______GUGUACCCCGU ..................((.((((.....))))))..............((((........))))(((((((...))))))).................. (-22.38 = -22.34 + -0.04)


| Location | 4,692,734 – 4,692,832 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.88 |
| Mean single sequence MFE | -31.05 |
| Consensus MFE | -20.31 |
| Energy contribution | -20.20 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4692734 98 - 22224390 GAAAUG--UGC----------CUCGCGUUAU-------UCGUUUCGCGGACCUGAA-AGGGCUGUAAAAAGGGAUAAAAUCAUAAGCCCAAAUUUAUCGCACUCGACAGUCGAGGCA--A ......--(((----------(((((.((((-------((.(((..(((.(((...-.))))))..)))..))))))...................(((....)))..).)))))))--. ( -27.10) >DroPse_CAF1 5904 109 - 1 GAAGUUGCUGCCGCUGCUGCUCUCGCGU--G-------CGGCAUCGCGAGGUUGA--AGGGCUGUAAAAAGGGAUAAAAUCAUAAGCCCAAAUUUAUCGCACUCGACAGCCAAGGCACAA ....(((.((((...((((...(((.((--(-------(((.(((....)))...--.(((((.....................))))).......)))))).)))))))...))))))) ( -35.00) >DroSim_CAF1 19263 106 - 1 GAAAUG--UGC----------CUCGCGUUUUUCGUUUUUCGUUUCGCGGGCCUGAAAAGGGCUGUAAAAAGGGAUAAAAUCAUAAGCCCAAAUUUAUCGCACUCGACAGUCGAGGCA--A ......--(((----------(((((.....(((.......(((..(((.(((....))).)))..))).(.((((((..............)))))).)...)))..).)))))))--. ( -29.14) >DroEre_CAF1 46576 98 - 1 GAAAUG--UGC----------CUCGCGUUUU-------UCGUUUCGCGAGCCUGAAAA-GGCUGUAAAAAGGGAUCAAAUCAUAAGCCCAAAUUUAUCGCACUCGACAGUCGAGGCA--A ......--(((----------(((((.....-------(((....((((((((....)-)))........(((.............))).......))))...)))..).)))))))--. ( -30.02) >DroYak_CAF1 43514 98 - 1 GAAAUG--UGC----------CUCGCGUUUU-------UCGUUUCGCGAGCCUGAAAA-GGCUGUAAAAAGGGAUAAAAUCAUAAGCCCAAAUUUAUCGCACUCGACAGUCGAGGCA--A ......--(((----------(((((.....-------(((....((((((((....)-)))........(((.............))).......))))...)))..).)))))))--. ( -30.02) >DroPer_CAF1 5085 109 - 1 GAAGUUGCUGCCGCUGCUGCUCUCGCGU--G-------CGGCAUCGCGAGGUUGA--AGGGCUGUAAAAAGGGAUAAAAUCAUAAGCCCAAAUUUAUCGCACUCGACAGCCAAGGCACAA ....(((.((((...((((...(((.((--(-------(((.(((....)))...--.(((((.....................))))).......)))))).)))))))...))))))) ( -35.00) >consensus GAAAUG__UGC__________CUCGCGUUUU_______UCGUUUCGCGAGCCUGAA_AGGGCUGUAAAAAGGGAUAAAAUCAUAAGCCCAAAUUUAUCGCACUCGACAGUCGAGGCA__A ........(((..........((((((.................)))))).........((((((.....(.((((((..............)))))).).....))))))...)))... (-20.31 = -20.20 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:22 2006