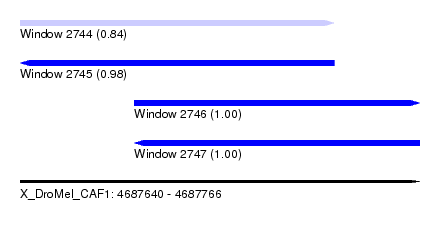
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,687,640 – 4,687,766 |
| Length | 126 |
| Max. P | 0.998373 |

| Location | 4,687,640 – 4,687,739 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 67.03 |
| Mean single sequence MFE | -22.95 |
| Consensus MFE | -11.48 |
| Energy contribution | -12.10 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4687640 99 + 22224390 UGAAAUUCCUUCACUG-UGGGGG-GGGGAGG-AAAUACUCCACCA--------AAAAAGCAUAAACAAAAAAAAAUGGCAAAAAUAUUCGCACACUUUGCGUGUGCG--------UGU .......(((((....-.)))))-((((((.-.....)))).)).--------...................................(((((((.....)))))))--------... ( -25.20) >DroSec_CAF1 41195 90 + 1 UGAAAUUCCUUCACUGCUGGAGU-GGGGAGC-AAAUACUCAUCCA--------AAAAAGCAUAAACAAAAAAAAA----------AUUCGCACACUUUGCGUGUGCG--------UGU ...........(((((((....(-((((((.-.....))).))))--------....))))..............----------....((((((.....)))))))--------)). ( -23.40) >DroEre_CAF1 41581 79 + 1 UCAAAUUCUUUCGCUG---GAGG----GGGGAAAAAGUUCCACCA----------------------AAA--AAAUGGCAAAAAUAUUCGCACACUUUGGGUUUGCG--------UGU ............((((---....----((((((....)))).)).----------------------...--...)))).........((((.((.....)).))))--------... ( -14.00) >DroYak_CAF1 38448 113 + 1 UGAAAUUCUUUCGCGG---GGGGGAAGGAGCAAAAAGCUCCACCAAACAAAAAAAAAAGUAUAAGCAAAA--AAAUGGCAAAAAUAUUCGCACACUUUGCGUGUGCGUGAGUGUGUGU .....((((..(....---)..))))(((((.....))))).......................((....--.....)).........(((((((.(..((....))..)))))))). ( -29.20) >consensus UGAAAUUCCUUCACUG___GAGG_GGGGAGC_AAAAACUCCACCA________AAAAAGCAUAAACAAAA__AAAUGGCAAAAAUAUUCGCACACUUUGCGUGUGCG________UGU .....................((...((((.......)))).))............................................(((((((.....)))))))........... (-11.48 = -12.10 + 0.62)



| Location | 4,687,640 – 4,687,739 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 67.03 |
| Mean single sequence MFE | -23.80 |
| Consensus MFE | -10.81 |
| Energy contribution | -12.50 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4687640 99 - 22224390 ACA--------CGCACACGCAAAGUGUGCGAAUAUUUUUGCCAUUUUUUUUUGUUUAUGCUUUUU--------UGGUGGAGUAUUU-CCUCCCC-CCCCCA-CAGUGAAGGAAUUUCA ...--------(((((((.....)))))))...................(((.(((((.......--------.((.((((.....-.))))))-......-..))))).)))..... ( -23.69) >DroSec_CAF1 41195 90 - 1 ACA--------CGCACACGCAAAGUGUGCGAAU----------UUUUUUUUUGUUUAUGCUUUUU--------UGGAUGAGUAUUU-GCUCCCC-ACUCCAGCAGUGAAGGAAUUUCA ...--------(((((((.....)))))))...----------......(((.((((((((....--------(((..((((....-)))).))-)....)))).)))).)))..... ( -25.00) >DroEre_CAF1 41581 79 - 1 ACA--------CGCAAACCCAAAGUGUGCGAAUAUUUUUGCCAUUU--UUU----------------------UGGUGGAACUUUUUCCCC----CCUC---CAGCGAAAGAAUUUGA ...--------(((......((((((.(((((....))))))))))--)..----------------------.((.((((....))))))----....---..)))........... ( -17.30) >DroYak_CAF1 38448 113 - 1 ACACACACUCACGCACACGCAAAGUGUGCGAAUAUUUUUGCCAUUU--UUUUGCUUAUACUUUUUUUUUUGUUUGGUGGAGCUUUUUGCUCCUUCCCCC---CCGCGAAAGAAUUUCA ...........(((((((.....)))))))............((((--((((((....((..........))..((.(((((.....)))))..))...---..)))))))))).... ( -29.20) >consensus ACA________CGCACACGCAAAGUGUGCGAAUAUUUUUGCCAUUU__UUUUGUUUAUGCUUUUU________UGGUGGAGCAUUU_CCUCCCC_CCCC___CAGCGAAAGAAUUUCA ...........(((((((.....)))))))...............................................(((((.....))))).......................... (-10.81 = -12.50 + 1.69)



| Location | 4,687,676 – 4,687,766 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 74.74 |
| Mean single sequence MFE | -19.56 |
| Consensus MFE | -16.17 |
| Energy contribution | -17.93 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.998373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4687676 90 + 22224390 CCACCA--------AAAAAGCAUAAACAAAAAAAAAUGGCAAAAAUAUUCGCACACUUUGCGUGUGCG--------UGUGUGGGGGAAAGUAAUUUACCAACACAG .(((..--------.....((.................))..........((((((.....)))))))--------))((((..(((((....))).))..)))). ( -20.33) >DroSec_CAF1 41232 80 + 1 CAUCCA--------AAAAAGCAUAAACAAAAAAAAA----------AUUCGCACACUUUGCGUGUGCG--------UGUGUGGGGGAAAGUAAUUUACCAACACAG ......--------......................----------...(((((((.....)))))))--------..((((..(((((....))).))..)))). ( -20.10) >DroEre_CAF1 41613 74 + 1 CCACCA----------------------AAA--AAAUGGCAAAAAUAUUCGCACACUUUGGGUUUGCG--------UGUGUGAGGGAAAGUAAUUUACCAACACAG ...(((----------------------...--...)))..........((((.((.....)).))))--------(((((...(((((....))).)).))))). ( -15.10) >DroYak_CAF1 38484 104 + 1 CCACCAAACAAAAAAAAAAGUAUAAGCAAAA--AAAUGGCAAAAAUAUUCGCACACUUUGCGUGUGCGUGAGUGUGUGUGUGAGGGAAAGUAAUUUACCAACACAG .(((((..((...............((....--.....)).........(((((((.....)))))))))..)).)))((((..(((((....))).))..)))). ( -22.70) >consensus CCACCA________AAAAAGCAUAAACAAAA__AAAUGGCAAAAAUAUUCGCACACUUUGCGUGUGCG________UGUGUGAGGGAAAGUAAUUUACCAACACAG .................................................(((((((.....)))))))........(((((..((.(((....))).)).))))). (-16.17 = -17.93 + 1.75)



| Location | 4,687,676 – 4,687,766 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 74.74 |
| Mean single sequence MFE | -20.16 |
| Consensus MFE | -16.48 |
| Energy contribution | -16.48 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.997183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4687676 90 - 22224390 CUGUGUUGGUAAAUUACUUUCCCCCACACA--------CGCACACGCAAAGUGUGCGAAUAUUUUUGCCAUUUUUUUUUGUUUAUGCUUUUU--------UGGUGG .(((((.((..((.....))..)).)))))--------(((((((.....))))))).......(..(((......................--------)))..) ( -22.25) >DroSec_CAF1 41232 80 - 1 CUGUGUUGGUAAAUUACUUUCCCCCACACA--------CGCACACGCAAAGUGUGCGAAU----------UUUUUUUUUGUUUAUGCUUUUU--------UGGAUG .(((((.((..((.....))..)).)))))--------(((((((.....)))))))...----------.........(((((........--------))))). ( -19.60) >DroEre_CAF1 41613 74 - 1 CUGUGUUGGUAAAUUACUUUCCCUCACACA--------CGCAAACCCAAAGUGUGCGAAUAUUUUUGCCAUUU--UUU----------------------UGGUGG .((((..((.(((....)))))..))))((--------(.((((...((((((.(((((....))))))))))--)))----------------------))))). ( -17.90) >DroYak_CAF1 38484 104 - 1 CUGUGUUGGUAAAUUACUUUCCCUCACACACACACUCACGCACACGCAAAGUGUGCGAAUAUUUUUGCCAUUU--UUUUGCUUAUACUUUUUUUUUUGUUUGGUGG .((((..((.(((....)))))..))))(((.((..(((((((((.....))))))).........((.....--....))...............))..))))). ( -20.90) >consensus CUGUGUUGGUAAAUUACUUUCCCCCACACA________CGCACACGCAAAGUGUGCGAAUAUUUUUGCCAUUU__UUUUGUUUAUGCUUUUU________UGGUGG .(((((.((..((.....))..)).)))))........(((((((.....)))))))................................................. (-16.48 = -16.48 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:17 2006