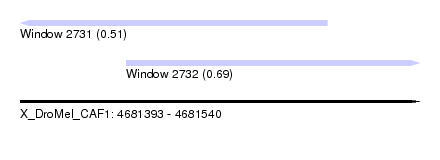
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,681,393 – 4,681,540 |
| Length | 147 |
| Max. P | 0.685009 |

| Location | 4,681,393 – 4,681,506 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.63 |
| Mean single sequence MFE | -23.71 |
| Consensus MFE | -16.79 |
| Energy contribution | -17.90 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4681393 113 - 22224390 CGAAAAUGGUUCAUCCAGUUGGCAAUUAUCUGUUUUCU------CCCCUAUGCCAAUGGAAACCAUGUCCAAGAGCAGUCAUUUUCCGAGGAAAACUCACCGCUUUUCGCACGCACACU .....((((((..((((.((((((..............------......))))))))))))))))(((.((((((.((..(((((....)))))...)).)))))).).))....... ( -26.45) >DroSec_CAF1 35077 119 - 1 CAAAGAUGGUUCAUCCAGUUGGCAAUUAUCUGUUAUUUCCAUUCCCCCUAUGCCAAUGGAAACCUUCUCCAAGAGCAGUCAUUUUCCGAGGAAAACCCACCGCUUUCCCCGCGCACACU ...(((.((((..((((.((((((..........................)))))))))))))).)))...(((((.((..(((((....)))))...)).)))))............. ( -23.37) >DroEre_CAF1 35415 107 - 1 CAAAAAUG-UUCAACCAGUUAGCAAUUAACUCUUAUUC------CCUCUAUGCCGAUGGAAACCAUGUCCGAGGGAAGUCAUUUUCCGAGGAAAACUCACCUC-UUCC----UAACACU ........-........(((((.............(((------((((...((..((((...))))))..)))))))..........((((........))))-...)----))))... ( -21.30) >consensus CAAAAAUGGUUCAUCCAGUUGGCAAUUAUCUGUUAUUU______CCCCUAUGCCAAUGGAAACCAUGUCCAAGAGCAGUCAUUUUCCGAGGAAAACUCACCGCUUUCC_C_CGCACACU .....((((((..((((.((((((..........................))))))))))))))))....((((((.((..(((((....)))))...)).))))))............ (-16.79 = -17.90 + 1.12)


| Location | 4,681,432 – 4,681,540 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.85 |
| Mean single sequence MFE | -30.65 |
| Consensus MFE | -19.40 |
| Energy contribution | -20.74 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.685009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4681432 108 + 22224390 GACUGCUCUUGGACAUGGUUUCCAUUGGCAUAGGGG------AGAAAACAGAUAAUUGCCAACUGGAUGAACCAUUUUCGUACACCUUGGGUAAAGAAUAAGCCAAGGGCAUAU ...(((((((((.(((((((((((((((((......------..............)))))).))))..))))))(((..(((.(....))))..)))...))))))))))... ( -32.75) >DroSec_CAF1 35116 114 + 1 GACUGCUCUUGGAGAAGGUUUCCAUUGGCAUAGGGGGAAUGGAAAUAACAGAUAAUUGCCAACUGGAUGAACCAUCUUUGUACACCUUGGGUAACCAAUAAACCAAGGGCAUAU ...((((((((((((.((((((((((((((.....(..((....))..).......)))))).))))..)))).))).........((((....))))....)))))))))... ( -36.60) >DroEre_CAF1 35449 107 + 1 GACUUCCCUCGGACAUGGUUUCCAUCGGCAUAGAGG------GAAUAAGAGUUAAUUGCUAACUGGUUGAA-CAUUUUUGUACACCUUGGGUAAUCAAUAAAUCAAAGAUAUAU ...(((((((.(..((((...))))...)...))))------))).....(((.((((((....(((.(.(-((....))).))))...)))))).)))............... ( -22.60) >consensus GACUGCUCUUGGACAUGGUUUCCAUUGGCAUAGGGG______AAAUAACAGAUAAUUGCCAACUGGAUGAACCAUUUUUGUACACCUUGGGUAAACAAUAAACCAAGGGCAUAU ...(((((((((....((((((((((((((..........................)))))).)))..))))).......(((.(....)))).........)))))))))... (-19.40 = -20.74 + 1.34)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:04 2006