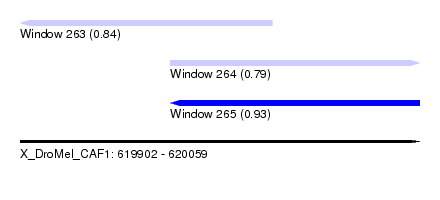
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 619,902 – 620,059 |
| Length | 157 |
| Max. P | 0.930004 |

| Location | 619,902 – 620,001 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.59 |
| Mean single sequence MFE | -22.83 |
| Consensus MFE | -21.08 |
| Energy contribution | -21.25 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
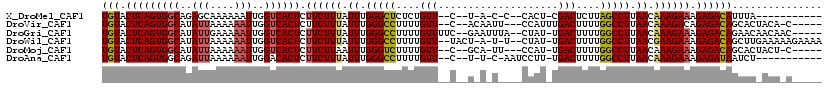
>X_DroMel_CAF1 619902 99 - 22224390 UGUACUCAGUGGCAGAGCAAAAAAUUGGUCACUCUUCUUUAUUUGGGCUCUCUGUU--C--U-A-C-C--CACU-CGACUCUUAGCCUUAACAAAGAAAGAGACAUUUA----------- (((.(((((((((.((........)).)))))).((((((.((.(((((....(((--.--.-.-.-.--....-.)))....))))).)).)))))).))))))....----------- ( -21.60) >DroVir_CAF1 4198 107 - 1 UGUACUCAGUGGCAUAUUAAAAAAUUGGUCACUCUUCUUUAUUUGGGCCUUUUGUU--C--ACAAUU---CCAUUUGACUUUUGGCCUUAACAAAGACAGAGACAGCACUACA-C----- (((.(((((((((..(((....)))..))))))..(((((.((.(((((....((.--.--.(((..---....)))))....))))).)).)))))..))))))........-.----- ( -22.60) >DroGri_CAF1 4149 110 - 1 UGUACUCAGUGGCAUAUUGAAAAAUUGGUCACUCUUCUUUAUUUGGGCCUUUUGUUUUC--GAAUUUA--CUAU-UGACUUUUGGCCUUAACAAAGAAAGAGACAGAACAACAAC----- (((.(((((((((..(((....)))..)))))).((((((.((.(((((....(((...--.......--....-.)))....))))).)).)))))).))))))..........----- ( -26.26) >DroWil_CAF1 5115 112 - 1 UGUACUCAGUGGCAUAUUAAAAAAUUGGUCACUCUUCUUUAUUUGGGCCUUUUGUU--UACU-A-U-U--CUAU-UGACUUUUGGCCUUAACGAAGAAAGAGACAGCUUGAAAAAGAAAA (((.(((((((((..(((....)))..)))))).(((((..((.(((((....(((--....-.-.-.--....-.)))....))))).))..))))).)))))).(((....))).... ( -26.80) >DroMoj_CAF1 3919 105 - 1 UGUACUCAGUGGCAUAUUAAAAAAUUGGUCACUCUUCUUAAUUUGGGUCUUUUGUU--C--GCA-UU---CCAU-UGACUUUUGGCCUUAACAAAGAAAGAGACAGCACUACU-C----- (((.(((((((((..(((....)))..)))))).(((((.....(((((....(((--.--...-..---....-.)))....))))).....))))).))))))........-.----- ( -19.10) >DroAna_CAF1 4393 101 - 1 UGUACUCAGUGGCAGAUUAAAAAAUUGGACACUCUUCUUUAUUUGGGCCUUUUGUU--C--U-U-C-AAUCCUU-UGACUUUUGGCCUUAACAAAGAAAGAGAUAAUCU----------- ....(((((((...((((....))))...)))).((((((.((.(((((.......--.--.-(-(-((....)-))).....))))).)).)))))).))).......----------- ( -20.64) >consensus UGUACUCAGUGGCAUAUUAAAAAAUUGGUCACUCUUCUUUAUUUGGGCCUUUUGUU__C__U_A_U____CCAU_UGACUUUUGGCCUUAACAAAGAAAGAGACAGCAC_ACA_C_____ (((.(((((((((..(((....)))..)))))).((((((.((.(((((....(((....................)))....))))).)).)))))).))))))............... (-21.08 = -21.25 + 0.17)
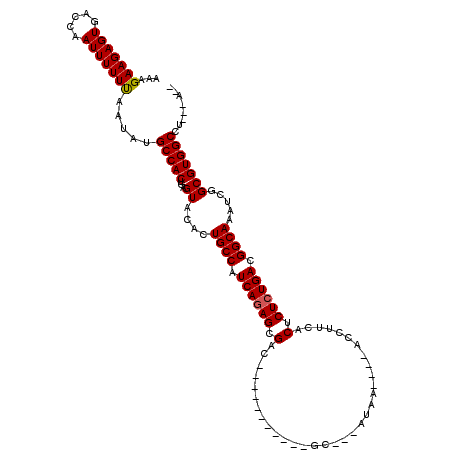
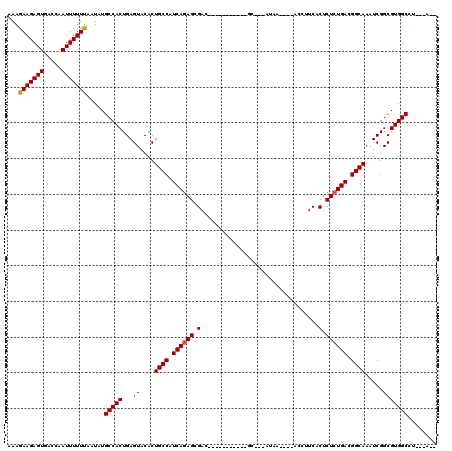
| Location | 619,961 – 620,059 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -29.47 |
| Consensus MFE | -22.11 |
| Energy contribution | -22.14 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 619961 98 + 22224390 AAAGAAGAGUGACCAAUUUUUUGCUCUGCCACUGAGUACACUGCCAUCAGAGCGAC-----------GC---AUAA----AGC-UCACUCUCUGACGGCAAAUCGGCGUGGCCU---GUA .....((((..(........)..))))(((((...((....((((.((((((.((.-----------((---....----.))-))...)))))).)))).....)))))))..---... ( -29.60) >DroVir_CAF1 4265 102 + 1 AAAGAAGAGUGACCAAUUUUUUAAUAUGCCACUGAGUACACUGCCAUCAGAGCGAC-----------ACGGCCAAAAG--GCCUUCACUCUCUGACGGCAAAUCGGCGUGGCUA---U-- ...(((((((.....))))))).....(((((...((....((((.((((((.(..-----------..((((....)--)))....).)))))).)))).....)))))))..---.-- ( -33.10) >DroGri_CAF1 4219 106 + 1 AAAGAAGAGUGACCAAUUUUUCAAUAUGCCACUGAGUACACUGCCAUCAGAGCGAC------CA---AUGGCUAAAAGAAGCCUUCACUCUCUGACGGCAAAUCGGCGUGGCCU---A-- ...(((((((.....))))))).....(((((...((....((((.((((((.(..------..---..((((......))))....).)))))).)))).....)))))))..---.-- ( -32.70) >DroWil_CAF1 5187 113 + 1 AAAGAAGAGUGACCAAUUUUUUAAUAUGCCACUGAGUACACUGCCAUCAAAGCGACAUGGACCAUUUGC---AUGG----ACCUUCACUCUCUGACGGCAAAUCGGCGUGGCCUUAUAUA ...(((((((.....))))))).....(((((...((....((((.(((.((.(....((.((((....---))))----.))....).)).))).)))).....)))))))........ ( -24.80) >DroMoj_CAF1 3984 101 + 1 UAAGAAGAGUGACCAAUUUUUUAAUAUGCCACUGAGUACACUGCCAUCAGAGCGAC-----------UA-UCCCAAGG--GAUUUCACUCUCUGACGGCAAAUCGGCGUGGCCU---A-- ...(((((((.....))))))).....(((((...((....((((.((((((.(..-----------.(-(((....)--)))....).)))))).)))).....)))))))..---.-- ( -30.50) >DroAna_CAF1 4454 97 + 1 AAAGAAGAGUGUCCAAUUUUUUAAUCUGCCACUGAGUACACUGCCAUCAGAGCGAC-----------GCC--AUGG----AGC-UCACUCUCUGACGGCAAAUCGGCGUGGCCU---A-- ...(((((((.....))))))).....(((((...((....((((.((((((.((.-----------((.--....----.))-))...)))))).)))).....)))))))..---.-- ( -26.10) >consensus AAAGAAGAGUGACCAAUUUUUUAAUAUGCCACUGAGUACACUGCCAUCAGAGCGAC___________GC___AUAA____ACCUUCACUCUCUGACGGCAAAUCGGCGUGGCCU___A__ ...(((((((.....))))))).....(((((...((....((((.((((((.(.................................).)))))).)))).....)))))))........ (-22.11 = -22.14 + 0.03)

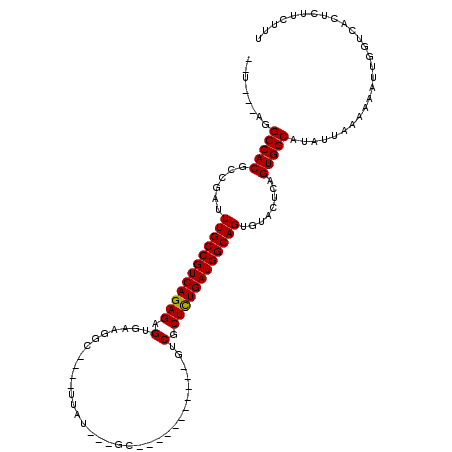
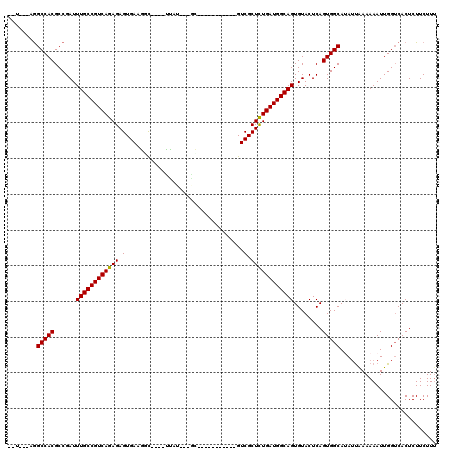
| Location | 619,961 – 620,059 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -36.07 |
| Consensus MFE | -26.79 |
| Energy contribution | -26.65 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
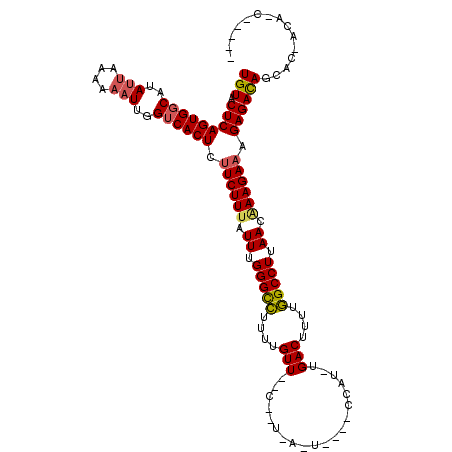
>X_DroMel_CAF1 619961 98 - 22224390 UAC---AGGCCACGCCGAUUUGCCGUCAGAGAGUGA-GCU----UUAU---GC-----------GUCGCUCUGAUGGCAGUGUACUCAGUGGCAGAGCAAAAAAUUGGUCACUCUUCUUU ...---..(((((......((((((((((((.(...-((.----....---))-----------..).))))))))))))........))))).((.(((....))).)).......... ( -35.64) >DroVir_CAF1 4265 102 - 1 --A---UAGCCACGCCGAUUUGCCGUCAGAGAGUGAAGGC--CUUUUGGCCGU-----------GUCGCUCUGAUGGCAGUGUACUCAGUGGCAUAUUAAAAAAUUGGUCACUCUUCUUU --.---..(((((......(((((((((((..((((.(((--(....))))..-----------.)))))))))))))))........)))))........................... ( -39.54) >DroGri_CAF1 4219 106 - 1 --U---AGGCCACGCCGAUUUGCCGUCAGAGAGUGAAGGCUUCUUUUAGCCAU---UG------GUCGCUCUGAUGGCAGUGUACUCAGUGGCAUAUUGAAAAAUUGGUCACUCUUCUUU --.---.(((((.((((..((((((((((((.((.((((((......)))).)---).------).).)))))))))))).........))))..(((....)))))))).......... ( -36.80) >DroWil_CAF1 5187 113 - 1 UAUAUAAGGCCACGCCGAUUUGCCGUCAGAGAGUGAAGGU----CCAU---GCAAAUGGUCCAUGUCGCUUUGAUGGCAGUGUACUCAGUGGCAUAUUAAAAAAUUGGUCACUCUUCUUU ........(((((......(((((((((..((((((.((.----((((---....)))).))...)))))))))))))))........)))))........................... ( -36.54) >DroMoj_CAF1 3984 101 - 1 --U---AGGCCACGCCGAUUUGCCGUCAGAGAGUGAAAUC--CCUUGGGA-UA-----------GUCGCUCUGAUGGCAGUGUACUCAGUGGCAUAUUAAAAAAUUGGUCACUCUUCUUA --.---..(((((......(((((((((((..((((.(((--(....)))-).-----------.)))))))))))))))........)))))........................... ( -35.14) >DroAna_CAF1 4454 97 - 1 --U---AGGCCACGCCGAUUUGCCGUCAGAGAGUGA-GCU----CCAU--GGC-----------GUCGCUCUGAUGGCAGUGUACUCAGUGGCAGAUUAAAAAAUUGGACACUCUUCUUU --.---..(((((......((((((((((((.(...-(((----....--)))-----------..).))))))))))))........)))))........................... ( -32.74) >consensus __U___AGGCCACGCCGAUUUGCCGUCAGAGAGUGAAGGC____UUAU___GC___________GUCGCUCUGAUGGCAGUGUACUCAGUGGCAUAUUAAAAAAUUGGUCACUCUUCUUU ........(((((......((((((((((((.(.................................).))))))))))))........)))))........................... (-26.79 = -26.65 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:48 2006