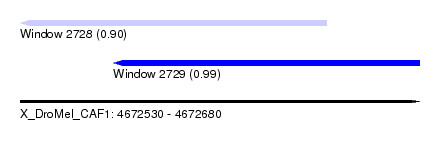
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,672,530 – 4,672,680 |
| Length | 150 |
| Max. P | 0.992800 |

| Location | 4,672,530 – 4,672,645 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -20.80 |
| Energy contribution | -20.30 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4672530 115 - 22224390 CUUGCUUCACUGGUUCCUUUGGUUGUCCGGCAACUAUUGUUGCUGGAUAUUUAGCAUUAGCUGGUGUCCGAUUUCUACUUUUUAACGA-----UUUAAGCUAGUUUAAAGUUGCAAAAGA .(((...(((..(((...((((.(((((((((((....))))))))))).))))....)))..)))..)))......(((((((((..-----.(((((....))))).)))).))))). ( -31.10) >DroSec_CAF1 33572 112 - 1 CUUGCUUCACUGGUUCCUUUGGUUGUCCGGCAACUAUUGUUGCUGGAUAUUUAGCAUUAGCUGGUGUCCGAUUUCUACUUUUUAACGA-----UUUGAACUA---UCAAGUUUCAGAAGA .(((...(((..(((...((((.(((((((((((....))))))))))).))))....)))..)))..)))......(((((....((-----(((((....---)))))))..))))). ( -31.30) >DroEre_CAF1 33974 114 - 1 UUUGCUUCACUGGUUCUUUUGGUUGUCCAGCAACUUUUGUUGUUGGGUAUUUAGCAUUAGUUCGUGCCCGGUUUCUACUUUUUAACGAUCUGAUUUCCACUC---GAAAUAUUAA---AA ..((((..((..(.....)..))(..((((((((....))))))))..)...))))((((.((((....(((....))).....)))).)))).........---..........---.. ( -23.90) >DroYak_CAF1 30766 112 - 1 CUCGCAUCACUGCUUCCAUUGGUUGUCCGGCAACUAUUGUUGCUCGAUAUUUAGCAUUAACUGGUGUCCGAUGUCGGCUUUUUAACGG-----UUUUCUCUU---GAAAUAUGGACAUGA ...(((....)))..........(((((((((((....))))))(((((((..(((((....)))))..)))))))...((((((.((-----....)).))---))))...)))))... ( -27.40) >consensus CUUGCUUCACUGGUUCCUUUGGUUGUCCGGCAACUAUUGUUGCUGGAUAUUUAGCAUUAGCUGGUGUCCGAUUUCUACUUUUUAACGA_____UUUAAACUA___GAAAGAUGCA_AAGA .(((...((((((((...((((.(((((((((((....))))))))))).))))....))))))))..)))................................................. (-20.80 = -20.30 + -0.50)


| Location | 4,672,565 – 4,672,680 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -26.41 |
| Consensus MFE | -20.80 |
| Energy contribution | -20.30 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4672565 115 - 22224390 UAGACAUUUCACCAUUUCUACUUA----CUCU-CACUUACCUUGCUUCACUGGUUCCUUUGGUUGUCCGGCAACUAUUGUUGCUGGAUAUUUAGCAUUAGCUGGUGUCCGAUUUCUACUU ((((.(((.((((...........----....-.....(((..(((.....)))......)))(((((((((((....)))))))))))...(((....)))))))...))).))))... ( -28.30) >DroSec_CAF1 33604 115 - 1 UAGAAAUUUCCCCAUUUCUACUUA----CCUC-CACUUACCUUGCUUCACUGGUUCCUUUGGUUGUCCGGCAACUAUUGUUGCUGGAUAUUUAGCAUUAGCUGGUGUCCGAUUUCUACUU ((((((((.(.((..........(----((..-.....(((..........)))......)))(((((((((((....)))))))))))...(((....))))).)...))))))))... ( -29.22) >DroEre_CAF1 34008 119 - 1 UAGACAUUUCCAC-UUUCCACUCAGCCACUACUUACAUAUUUUGCUUCACUGGUUCUUUUGGUUGUCCAGCAACUUUUGUUGUUGGGUAUUUAGCAUUAGUUCGUGCCCGGUUUCUACUU ..((((...(((.-...(((...(((.................)))....)))......))).)))).((((((....))))))((((((..(((....))).))))))........... ( -21.83) >DroYak_CAF1 30798 111 - 1 UAGACAUUUCUAU-UUACC-------CAUUGC-UACUUACCUCGCAUCACUGCUUCCAUUGGUUGUCCGGCAACUAUUGUUGCUCGAUAUUUAGCAUUAACUGGUGUCCGAUGUCGGCUU ..((((((.....-.((((-------...(((-((........(((....)))..........((((.((((((....)))))).))))..)))))......))))...))))))..... ( -26.30) >consensus UAGACAUUUCCAC_UUUCCACUUA____CUAC_CACUUACCUUGCUUCACUGGUUCCUUUGGUUGUCCGGCAACUAUUGUUGCUGGAUAUUUAGCAUUAGCUGGUGUCCGAUUUCUACUU .........................................(((...((((((((...((((.(((((((((((....))))))))))).))))....))))))))..)))......... (-20.80 = -20.30 + -0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:16:01 2006