
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,672,201 – 4,672,480 |
| Length | 279 |
| Max. P | 0.697403 |

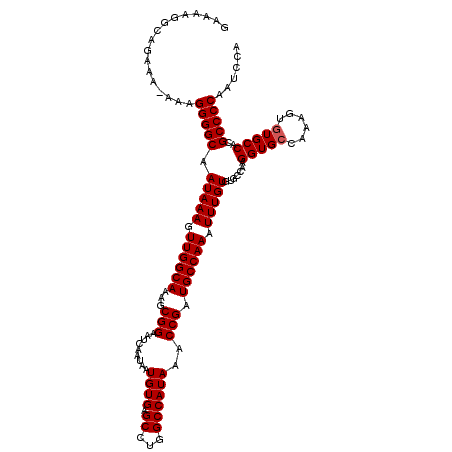
| Location | 4,672,201 – 4,672,320 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.51 |
| Mean single sequence MFE | -33.07 |
| Consensus MFE | -29.88 |
| Energy contribution | -30.12 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4672201 119 - 22224390 GAAAAGGCAGAAA-AAAGGGGCAAUAAAGUUGGCAAAGCGGAAUCAAUAAUGUGAGCCUGGCCAUAAACCGUUGCCAAAUUUGUGUGCCAAGGUGCCAAAGUUUGCCACGCCCCGAUCCA .............-...(((((.(((((.((((((..((((..(((......)))((...))......)))))))))).)))))(((.((((.(.....).)))).))))))))...... ( -32.90) >DroSec_CAF1 33213 119 - 1 GAAAAGGCAGAAA-AAAGGGGCAAUAAAGUUGGCAAAGCGGAAUCAAUAAUGUGAGCCCGGCCAUAAACCGUUGCCAAAUUUGUGUGCCAAGGUGCCAAAGUGUGCCACGCCCCAAUCCA .............-...(((((.......((((((..((((..(((......)))..))(((.((.....)).)))......)).))))))((..(......)..))..)))))...... ( -34.60) >DroEre_CAF1 33644 119 - 1 GAAAAGGCAGAAA-AAAGGGGCAAUAAAGUUGGCAAAGCGGAAUCAAUAAUGUGAGCCUGGCCAUAAACCGAUGCCAAAUUUGUGUGCCAAGGUGCCAAAGUGUGCCACGCCCCAAUCCA .............-...(((((.......(((((.........(((......)))(((((((((((((...........)))))).))).))))))))).(((...))))))))...... ( -33.10) >DroYak_CAF1 30406 120 - 1 GGAAAGGCAGAAAAAAAGGGGCAAUAAAGUUGGCAAAGCGGAAUCAAUAAUGUGUGCCUGGCCAUAAACCGAUGCCAAAUUUGUGUGUCAAGGUGCCAAAGUGUGCCACGCCCCAAUCCA .................(((((.(((((.((((((...(((.........((((.((...))))))..))).)))))).))))).......((..(......)..))..)))))...... ( -31.70) >consensus GAAAAGGCAGAAA_AAAGGGGCAAUAAAGUUGGCAAAGCGGAAUCAAUAAUGUGAGCCUGGCCAUAAACCGAUGCCAAAUUUGUGUGCCAAGGUGCCAAAGUGUGCCACGCCCCAAUCCA .................(((((.(((((.((((((...(((.........((((.((...))))))..))).)))))).))))).......(((((......)))))..)))))...... (-29.88 = -30.12 + 0.25)


| Location | 4,672,241 – 4,672,360 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.51 |
| Mean single sequence MFE | -31.32 |
| Consensus MFE | -30.12 |
| Energy contribution | -30.00 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4672241 119 - 22224390 CCGGUGUAUCUGUUCGGGUCAUAAAAACAGUGAUGCCCCAGAAAAGGCAGAAA-AAAGGGGCAAUAAAGUUGGCAAAGCGGAAUCAAUAAUGUGAGCCUGGCCAUAAACCGUUGCCAAAU .((((.....((..(((((((((.......(((((((((..............-...)))))......(((.....)))...))))....))))).))))..))...))))......... ( -28.23) >DroSec_CAF1 33253 119 - 1 CCGGUGUAUCUGUUCGGGCCAUAAAAACAGUGAUGCCCCAGAAAAGGCAGAAA-AAAGGGGCAAUAAAGUUGGCAAAGCGGAAUCAAUAAUGUGAGCCCGGCCAUAAACCGUUGCCAAAU .((((.....((..(((((((((.......(((((((((..............-...)))))......(((.....)))...))))....)))).)))))..))...))))......... ( -32.53) >DroEre_CAF1 33684 119 - 1 CCGGUGUAUCUGUUCGGGCCAUAAAAACAGUGACGCCCCAGAAAAGGCAGAAA-AAAGGGGCAAUAAAGUUGGCAAAGCGGAAUCAAUAAUGUGAGCCUGGCCAUAAACCGAUGCCAAAU ...(.(((((.(((..(((((.....(((.(((.(((((..............-...)))))......(((.....)))....)))....))).....)))))...))).)))))).... ( -30.83) >DroYak_CAF1 30446 120 - 1 ACGGUGUAUCUGUUCGGGCCAUAAAAACAGUGAUGCCCCAGGAAAGGCAGAAAAAAAGGGGCAAUAAAGUUGGCAAAGCGGAAUCAAUAAUGUGUGCCUGGCCAUAAACCGAUGCCAAAU ...(.(((((.(((..(((((....(((..((.((((((..................)))))).))..)))((((..(((..........))).)))))))))...))).)))))).... ( -33.67) >consensus CCGGUGUAUCUGUUCGGGCCAUAAAAACAGUGAUGCCCCAGAAAAGGCAGAAA_AAAGGGGCAAUAAAGUUGGCAAAGCGGAAUCAAUAAUGUGAGCCUGGCCAUAAACCGAUGCCAAAU .((((.....((..(((((((((.......(((((((((..................)))))......(((.....)))...))))....)))).)))))..))...))))......... (-30.12 = -30.00 + -0.13)



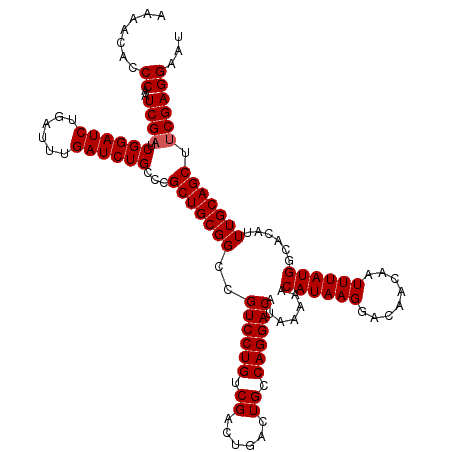
| Location | 4,672,360 – 4,672,480 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.61 |
| Mean single sequence MFE | -34.42 |
| Consensus MFE | -33.05 |
| Energy contribution | -33.30 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4672360 120 - 22224390 AAAACACCCAAUCGAUCGGAUCUGAUUUGAUCUGCCCGCUGCGGCCGUCCUGUCGACUGACUGCCAGGACAUAAAAACAUAAGGACAACAAUUUAUGGCACAUUUGCAGCUUCGAGGAAU .......((..((((.((((((......))))))...(((((((..((((((.((......)).)))))).......((((((........))))))......))))))).))))))... ( -34.10) >DroSec_CAF1 33372 120 - 1 AAAACUCCCAAUCGAUCGGAUCUGAUUUGAUCUGCCCGCUGCGGCCGUCCUGUCGACUGACUGCCAGGACAUAAAAACAUAAGGACAACAAUUUAUGGCACAUUUGCAGCUCCGAGGAAU .....(((...(((..((((((......))))))...(((((((..((((((.((......)).)))))).......((((((........))))))......)))))))..)))))).. ( -33.80) >DroEre_CAF1 33803 120 - 1 AAAACACCCAAUCGAUCGGAUCUGAUUUGAUCUGCCCGCUGCGGCCGUCCUGUCGACUGACUGCCAGGACAUAAAAACAUAAGGACAACAAUUUAUGGCACAUUUGCAGCUUCGAGGAAU .......((..((((.((((((......))))))...(((((((..((((((.((......)).)))))).......((((((........))))))......))))))).))))))... ( -34.10) >DroYak_CAF1 30566 120 - 1 AAAACUCCCCAUCGAUCGGAUCUGAUUUGAUCUGCCCGCUGCGGCCGUCCUGUCGACUGACUGCCAGGACAUAAAAACAUAAGGACAACAAUUUAUGGCACAUUUGCAGCUUCGAGGAAU ........((.((((.((((((......))))))...(((((((..((((((.((......)).)))))).......((((((........))))))......))))))).))))))... ( -35.70) >consensus AAAACACCCAAUCGAUCGGAUCUGAUUUGAUCUGCCCGCUGCGGCCGUCCUGUCGACUGACUGCCAGGACAUAAAAACAUAAGGACAACAAUUUAUGGCACAUUUGCAGCUUCGAGGAAU .......((..((((.((((((......))))))...(((((((..((((((.((......)).)))))).......((((((........))))))......))))))).))))))... (-33.05 = -33.30 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:15:59 2006